This site deals with Molecular Genetics and Biochemistry, particularly in relation to genetic mechanisms of biological evolution. The site is searchable using the "Search this blog" box at top left. Blue terms hyperlink to explanatory items so that you may navigate through items providing more detail. The Molecular Genetics Overview and Biochemistry Overview lead into the items. Use the "back" function to return to each departure item.
The “Guide-Glossary” link below each item provides a glossary of terms ( # > 0 ), as a pop-up when reading within a Section, or as sub-script when visiting an Item.
12/31/2007
Molecular Genetics Overview
Molecular genetics is the study of molecules and mechanisms involved in genetic inheritance. Archival information molecules are long polymers of deoxyribonucleic acid (DNA) comprising bases in specific sequence. The bases adenine (A), thymine (T), cytosine (C), and guanine (G), function as codon triplets – sequences of three bases that code for specific amino acids or for translation initiation (start codons) or termination (stop codons). Uracil is substituted for thymine in RNA.
The segments of DNA that contain protein-coding instructions are called genes, and these gene sequences comprise a portion of the total genome of a cell. The genome includes both the genes (coding-sequences, domains) and the non-coding sequences – both exons, which include open reading frames, and introns.
Because the 64 possible combinations of G-A-T-C code for only the 20 amino acids commonly found in proteins, the code is 'degenerate' (redundant) with more than one triplet combination coding for each amino acid. (This code reduncancy provides hereditary stability by reducing mutation errors.) The double helix of DNA comprises paired nucleotide strands in which the nucleobases are hydrogen bonded to complementary bases in the adjacent chain. Adenine pairs with thymine or uracil (A-TU), and cytosine pairs with guanine (CG).
During cellular reproduction, strands of archival DNA are copied or replicated. Transcription is the first step in gene expression – DNA instructions are converted into mRNA codons, rRNAs, miRNAs, and tRNAs. Coding instructions of nucleotide sequences in archival DNA, which have been transcribed and processed into mRNAs are then translated into polypeptides and proteins at cytoplasmic ribosomes. Translation is the ultimate step in gene expression, in which archival genetic instructions are converted into specified sequences of amino acids in peptides, polypeptides, and proteins.
In prokaryotic cells – without a nuclear membrane – translation into polypeptides and proteins may begin prior to termination of transcription. The molecular genetics of eukaryotic cells is more complicated than that of prokaryotes. Various molecules of ribonucleic acid (RNA) participate in the transcription of the DNA code into processed mRNA in a series of RNA processing stages including capping, polyadenylation, and pre-mRNA splicing.
Following pre-mRNA processing, RNAs undergo extranuclear transfer. Mature RNAs may undergo post-transcriptional modulation (via miRNAs) before translation of the archival DNA instructions into specific sequences of amino acids in the polypeptides and proteins that participate in cellular function and structure. Transfer RNAs (tRNA) deliver specific amino acids to the cytoplasmic ribosomes along the rough endoplasmic reticulum. Ribosomal RNAs participate in assembly of polypeptides and proteins at ribosomes. Here RNAs serve as ribozymes – non-proteinaceous enzymes.
A number of processes are involved in control of cellular function through the maintenance of accuracy of genetic inheritance – damage to DNA is repaired, and faulty RNA is destroyed by nonsense-mediated decay or nonstop decay.
DNA damage may result from replication errors, incorporation of mismatched nucleotides (substitution errors – transitions and transversions), damage by oxygen radicals, hydroxyl radicals, ionizing or ultraviolet radiation, toxins, alkylating agents, and chemotherapy agents. A number of vital mechanisms repair DNA damage to bases (including C to T, C to U, and T U mismatch) and to strands, including double strand breaks. All organisms, prokaryotic and eukaryotic, utilize at least three enzymatic excision-repair mechanisms for damaged bases: base excision repair, mismatch repair, and nucleotide excision repair.
Given the importance of mRNA as an information-carrying molecule, faulty pre-mRNAs and mRNAs must be eliminated – they are destroyed by nonsense-mediated decay or nonstop decay:
1. A pre-mRNA made from a mutant gene usually has an exon junction complex (EJC) in the wrong position. This error activates nonsense-mediated decay (NMD) and destroys the pre-mRNA before it can be used to make flawed proteins. There are at least two kinds of NMD: one requires the protein UPF2 and the other does not.
2. Nonstop decay is mRNA turnover mechanism that has none of the properties of normal mRNA turnover or of NMD. A multi-enzyme complex called the exosome is important for nonstop decay. The exosome is the site for binding of a specific adapter protein called Ski7p. Nonstop decay shares none of the enzymes required for nonsense-mediated decay.
Just as cells repair DNA, they must also maintain the proteome by managing damaged proteins. Heat stress denaturates proteins, causing weakening of polar bonds and exposure of hydrophobic groups. The cellular stress response (heat-shock response) protects organisms from damage resulting from environmental stressors such as heat, UV light, trace metals, and xenobiotics. Stress genes are activated to rapidly synthesize stress proteins, which are highly conserved in biological evolution and play similar roles in organisms from bacteria to humans. Normally, several constitutive stress proteins are present at low levels to function as molecular chaperones, so as to facilitate folding, assembly, and distribution of newly synthesized proteins. For the environmentally stressed cell, stress proteins protect and repair vulnerable protein targets, and play a role in the lysosomal and ubiquitin protein degradation pathways (for removal of unsalvageable proteins). Thus, the cellular stress response performs orchestrated induction of key proteins necessary for cellular protein repair and degradation systems.
Topics: item links : A : adhesion molecules : apoptosis : C : cell biology : chromosomes : D : damage/repair DNA : decay : E : energy : enzymes, specific enzymes : evo-devo : evolution : G : genome : M : mechanisms of evolution : metabolism : molecular genetics : mutation : P : photosynthesis : processing RNA : proteins, specific proteins : R : regulation · cellular differentiation . gene regulation . metabolic regulation : replication : repair/damage DNA : research : RNA : S : serial endosymbiosis theory : signaling : splicing : T : tables : transcription : translation :
▲ Top ▲
The segments of DNA that contain protein-coding instructions are called genes, and these gene sequences comprise a portion of the total genome of a cell. The genome includes both the genes (coding-sequences, domains) and the non-coding sequences – both exons, which include open reading frames, and introns.
Because the 64 possible combinations of G-A-T-C code for only the 20 amino acids commonly found in proteins, the code is 'degenerate' (redundant) with more than one triplet combination coding for each amino acid. (This code reduncancy provides hereditary stability by reducing mutation errors.) The double helix of DNA comprises paired nucleotide strands in which the nucleobases are hydrogen bonded to complementary bases in the adjacent chain. Adenine pairs with thymine or uracil (A-TU), and cytosine pairs with guanine (CG).
During cellular reproduction, strands of archival DNA are copied or replicated. Transcription is the first step in gene expression – DNA instructions are converted into mRNA codons, rRNAs, miRNAs, and tRNAs. Coding instructions of nucleotide sequences in archival DNA, which have been transcribed and processed into mRNAs are then translated into polypeptides and proteins at cytoplasmic ribosomes. Translation is the ultimate step in gene expression, in which archival genetic instructions are converted into specified sequences of amino acids in peptides, polypeptides, and proteins.
In prokaryotic cells – without a nuclear membrane – translation into polypeptides and proteins may begin prior to termination of transcription. The molecular genetics of eukaryotic cells is more complicated than that of prokaryotes. Various molecules of ribonucleic acid (RNA) participate in the transcription of the DNA code into processed mRNA in a series of RNA processing stages including capping, polyadenylation, and pre-mRNA splicing.
Following pre-mRNA processing, RNAs undergo extranuclear transfer. Mature RNAs may undergo post-transcriptional modulation (via miRNAs) before translation of the archival DNA instructions into specific sequences of amino acids in the polypeptides and proteins that participate in cellular function and structure. Transfer RNAs (tRNA) deliver specific amino acids to the cytoplasmic ribosomes along the rough endoplasmic reticulum. Ribosomal RNAs participate in assembly of polypeptides and proteins at ribosomes. Here RNAs serve as ribozymes – non-proteinaceous enzymes.
A number of processes are involved in control of cellular function through the maintenance of accuracy of genetic inheritance – damage to DNA is repaired, and faulty RNA is destroyed by nonsense-mediated decay or nonstop decay.
DNA damage may result from replication errors, incorporation of mismatched nucleotides (substitution errors – transitions and transversions), damage by oxygen radicals, hydroxyl radicals, ionizing or ultraviolet radiation, toxins, alkylating agents, and chemotherapy agents. A number of vital mechanisms repair DNA damage to bases (including C to T, C to U, and T U mismatch) and to strands, including double strand breaks. All organisms, prokaryotic and eukaryotic, utilize at least three enzymatic excision-repair mechanisms for damaged bases: base excision repair, mismatch repair, and nucleotide excision repair.
Given the importance of mRNA as an information-carrying molecule, faulty pre-mRNAs and mRNAs must be eliminated – they are destroyed by nonsense-mediated decay or nonstop decay:
1. A pre-mRNA made from a mutant gene usually has an exon junction complex (EJC) in the wrong position. This error activates nonsense-mediated decay (NMD) and destroys the pre-mRNA before it can be used to make flawed proteins. There are at least two kinds of NMD: one requires the protein UPF2 and the other does not.
2. Nonstop decay is mRNA turnover mechanism that has none of the properties of normal mRNA turnover or of NMD. A multi-enzyme complex called the exosome is important for nonstop decay. The exosome is the site for binding of a specific adapter protein called Ski7p. Nonstop decay shares none of the enzymes required for nonsense-mediated decay.
Just as cells repair DNA, they must also maintain the proteome by managing damaged proteins. Heat stress denaturates proteins, causing weakening of polar bonds and exposure of hydrophobic groups. The cellular stress response (heat-shock response) protects organisms from damage resulting from environmental stressors such as heat, UV light, trace metals, and xenobiotics. Stress genes are activated to rapidly synthesize stress proteins, which are highly conserved in biological evolution and play similar roles in organisms from bacteria to humans. Normally, several constitutive stress proteins are present at low levels to function as molecular chaperones, so as to facilitate folding, assembly, and distribution of newly synthesized proteins. For the environmentally stressed cell, stress proteins protect and repair vulnerable protein targets, and play a role in the lysosomal and ubiquitin protein degradation pathways (for removal of unsalvageable proteins). Thus, the cellular stress response performs orchestrated induction of key proteins necessary for cellular protein repair and degradation systems.
Topics: item links : A : adhesion molecules : apoptosis : C : cell biology : chromosomes : D : damage/repair DNA : decay : E : energy : enzymes, specific enzymes : evo-devo : evolution : G : genome : M : mechanisms of evolution : metabolism : molecular genetics : mutation : P : photosynthesis : processing RNA : proteins, specific proteins : R : regulation · cellular differentiation . gene regulation . metabolic regulation : replication : repair/damage DNA : research : RNA : S : serial endosymbiosis theory : signaling : splicing : T : tables : transcription : translation :
▲ Top ▲
12/24/2007
actin binding protein
Actin binding proteins (ABP) function in:
● sequestration of actin monomers
● severing actin microfilaments
● capping of actin microfilaments
● cross-linking of microfilaments
● cell motility
[m]
Џ Harvard video of actin-myosin action : beautiful Flash 8 animation - Inner Life of the Cell, which shows gelsolin severing and capping an actin filament ; and Interpretation: Inner Life of the Cell Џ
● sequestration of actin monomers
● severing actin microfilaments
● capping of actin microfilaments
● cross-linking of microfilaments
● cell motility
[m]
Џ Harvard video of actin-myosin action : beautiful Flash 8 animation - Inner Life of the Cell, which shows gelsolin severing and capping an actin filament ; and Interpretation: Inner Life of the Cell Џ
activator
In molecular genetics, an activator is a DNA-binding protein that regulates one or more genes by increasing the rate of transcription. Coactivators act in concert with other transcription factors. CREB binding protein (CBP) is a coactivator-transcription factor that binds with the cAMP response element (CRE).
Activators combine with a gene sequence to up-regulate gene expression. A catabolite gene activator protein (im), also called catabolite gene activator, is a cAMP dependent RNA polymerase that up-regulates gene expression by binding the polymerase at or near the DNA to be transcribed (families). In developmental biology or embryology, activators are substances that stimulate the development of a particular embryonic structure (inductor and organizer).
In biochemistry, an activator is a substance that combines with an enzyme to increase its catalytic activity.
In microbiology, autoinducers are signaling molecules in bacterial quorum sensing that regulate mRNA production for specific genes in response to population density. Autoinducer molecules, which include certain peptides and compounds known as homoserine lactones, can interact with specific repressor or enhancer sequences in DNA. The presence or absence of the autoinducer controls the production of mRNA, and therefore regulates gene expression into protein.
▲ Top ▲
Activators combine with a gene sequence to up-regulate gene expression. A catabolite gene activator protein (im), also called catabolite gene activator, is a cAMP dependent RNA polymerase that up-regulates gene expression by binding the polymerase at or near the DNA to be transcribed (families). In developmental biology or embryology, activators are substances that stimulate the development of a particular embryonic structure (inductor and organizer).
In biochemistry, an activator is a substance that combines with an enzyme to increase its catalytic activity.
In microbiology, autoinducers are signaling molecules in bacterial quorum sensing that regulate mRNA production for specific genes in response to population density. Autoinducer molecules, which include certain peptides and compounds known as homoserine lactones, can interact with specific repressor or enhancer sequences in DNA. The presence or absence of the autoinducer controls the production of mRNA, and therefore regulates gene expression into protein.
▲ Top ▲
adenylate cyclases
Adenylyl (adenylate) cyclases are transmembrane enzymes that cross the membrane twelve times, and which convert ATP to the second-messenger cAMP (3',5' cyclic AMP) and pyrophosphate (Pi). Likewise, guanylate cyclases convert GTP to the second messenger, cGMP.
Adenylyl cyclases are coincidence detectors, meaning that they are only activated by several different signals occurring together – they are modulated by G-proteins, forskolin, Ca2+/calmodulin, and other class-specific substrates.
Glossary : A . B . C . D . E . F . G . H . I . J . K . L . M . N . O . P . Q . R . S . T . U . V . W . X . Y . Z .
Gray Sites • Abiogenesis & Evolution • Algorithms of Evolution • Cell Biology • Chemistry of Life • Cyanobacteria • Enzymes • Evo Devo • Immunology • Mechanisms of Evolution • Molecular Biology • Origin of Life • Paleogeology • Refuting ID • Serial Endosymbiosis • Stromatolites • Taxonomy Phylogeny • Virus • White Sites • Diagrams & Tables • Eubacteria & Archaea • Molecule • Molecular Paths • Pathways • Photosynthesis • Black Sites • Endosymbiosis • Organics •
Adenylyl cyclases are coincidence detectors, meaning that they are only activated by several different signals occurring together – they are modulated by G-proteins, forskolin, Ca2+/calmodulin, and other class-specific substrates.
Glossary : A . B . C . D . E . F . G . H . I . J . K . L . M . N . O . P . Q . R . S . T . U . V . W . X . Y . Z .
Gray Sites • Abiogenesis & Evolution • Algorithms of Evolution • Cell Biology • Chemistry of Life • Cyanobacteria • Enzymes • Evo Devo • Immunology • Mechanisms of Evolution • Molecular Biology • Origin of Life • Paleogeology • Refuting ID • Serial Endosymbiosis • Stromatolites • Taxonomy Phylogeny • Virus • White Sites • Diagrams & Tables • Eubacteria & Archaea • Molecule • Molecular Paths • Pathways • Photosynthesis • Black Sites • Endosymbiosis • Organics •
adhesion molecules
Cell adhesion relies upon specialized transmembrane adhesion proteins, cell adhesion molecules (CAMs), through which cell-cell and cell-matrix interactions are mediated. The molecules of these families of adhesion molecules act through receptor-ligand interactions that usually extend from the intracellular space to the extracellular space where they may bind to other cell membranes (cell-cell) or to the extracellular matrix (cell-matrix, cell-ECM).
Tables Cell Adhesion Molecules Cell signaling Immune Cytokines Second Messengers
▼ cadherins : cadherin superfamily : catenins : cell-ECM : E-cadherin : FAK : families of CAMs : ICAM : immunoglobulin superfamily : importance of cell adhesion : integrins : selectins : signaling ▼
Cell adhesion is important in:
1. maintaining contact within solid tissue
2. embryogenesis (morphogenesis)
3. migration of single cells such as leukocytes within multicellular organisms
4. maintaining contact between neuronal elements
5. virulence of virions and bacteria
There are several families of adhesion proteins, each with specific ligands:
1. integrins with heterophilic attachments to different (hetero) ligands, including selectins with heterophilic attachments to carbohydrate ligands,
2. selectins with heterophilic attachments to carbohydrate ligands
3. Ig superfamily proteins with heterophilic attachments to: a) integrin ligands, and b) Ig superfamily proteins of a different (hetero) type, and homophilic attachments to the Ig superfamily proteins of the same (homo) type
4. cadherins with homophilic attachments to cadherins of the same type.
Cell-extracellular matrix interactions play a central role in tissue architecture and turnover. In particular, integrin-mediated cell adhesion participates in biochemical and physical signals. Cell adhesion molecules of the immunoglobulin and cadherin families, integrin heterodimers, and extracellular matrix glycoproteins, are widely expressed in the nervous system. Adhesion molecules of the immunoglobulin superfamily are involved in cell-cell adhesion, especially important during embryogenesis, wound healing, and the inflammatory response. Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion proteins. Integrins are heterodimeric molecules that function both as cell-substratum and cell-cell adhesion receptors. LEC-CAMs are cell adhesion molecules with lectin-like domains that mediate white blood cell/endothelial cell adhesion in the leukocyte adhesion cascade. Homing receptors target lymphocytes to specific lymphoid tissue.
Engagement of ICAM-1, a member of the immunoglobulin supergene family, has been documented to activate specific kinases through phosphorylation, resulting in activation of transcription factors, increased cytokine production, increased cell membrane protein expression, production of reactive oxygen species, and cell proliferation.
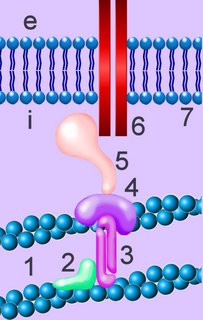 Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion proteins (right - click to enlarge). The classic cadherins are defined by a conserved intracellular (i) domain which mediates interactions with cytoplasmic proteins termed catenins: α- and β-catenin. β-catenin (5) binds to both the C-terminus of the cadherin intracellular domain (6) and the N-terminus of α-catenin (4). α-catenin binds to a number of proteins involved in actin binding, bundling and polymerisation, as well as binding directly to F-actin (1) of the cytoskeleton, here through α-actin (3) in association with vinculin (2).
Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion proteins (right - click to enlarge). The classic cadherins are defined by a conserved intracellular (i) domain which mediates interactions with cytoplasmic proteins termed catenins: α- and β-catenin. β-catenin (5) binds to both the C-terminus of the cadherin intracellular domain (6) and the N-terminus of α-catenin (4). α-catenin binds to a number of proteins involved in actin binding, bundling and polymerisation, as well as binding directly to F-actin (1) of the cytoskeleton, here through α-actin (3) in association with vinculin (2).
Absence of α- or β-catenin results in defective cell adhesion and failure of cadherin-catenin complexes to associate with the actin cytoskeleton.
Members of the cadherin superfamily are grouped according to the presence of one or more cadherin repeats in their extracellular (e) domains. Arrays of these (approx. 110 residue) domains form the intermolecular surfaces that mediate cadherin-mediated cell-cell interactions. Patel et al., 2003). The extracellular domains of type I and II (chordate) cadherins consist of five cadherin repeats (CRs). Type III (invertebrate and non-mammalian vertebrate) cadherins have variable numbers of CRs and also contain a region termed the primitive classical cadherin domain (PCCD) which, together with variable numbers of EGF-like and laminin G repeats, lies between the CRs and the transmembrane helix.
E-cadherin is the prototypic member of the cadherin transmembrane protein family, and regulates cell-to-cell adhesion by interacting with (homotype) E-cadherin molecules on opposing cell surfaces. E-cadherin's function in cell adhesion is also critically dependent on its ability to interact, through its cytoplasmic domain, with catenin proteins. A diverse collection of defects alter cadherin-catenin function in cancer cells.
FAK:
Some adhesion-mediated events include activation of focal adhesion kinase (FAK) with subsequent alterations in the cytoskeleton and cell morphology, changes in adhesion strength, and changes in cellular responsiveness to mechanical stimuli.[5] Fibronectin and type I collagen binding requires transcription factors (c-Fos and c-Jun) which are important in cell proliferation. A signaling pathway involving the phosphorylation of focal adhesion kinase and mitogen-activated kinases has also shown to be transiently increased in osteoblasts on fibronectin and type I collagen, but not in cells on poly-L-lysine.[6]
Ig superfamily:
The immunoglobulin superfamily is evolutionarily ancient, is widely expressed, and is constitutive or long-term up-regulated. Immunoglobulin antibodies are released by activated B cells of the immune system, on which they also act as marker, surface proteins.
Integrins:
Integrins are heterodimeric proteins so named because they integrate the function of the cell with the outside world.
Selectins:
Single transmembrane polypeptide selectins occur only in vertebrates, where they undergo calcium dependent binding via their amino-terminal domain to saccharides. The selectin family mediates function/migration of circulatory cells and the leukocyte adhesion cascade: L-leukocytes; E-endothelial cells; P-platelets and endothelial cells. The family possess large, highly glycosylated, extracellular domains, a single spanning transmembrane domain, and a small cytoplasmic tail. Selectins are rapidly downregulated by proteolytic cleavage.
Signaling molecules:
Some signaling molecules act as adhesion receptors and cluster in focal adhesions upon ligand binding. (Rho protein). Adhesion receptors promote the assembly of multi-molecular, intracellular complexes providing mechanical stability and generating intracellular signals that influence cell behavior. The functional properties of adhesion receptor complexes include roles in cell and tissue morphogenesis, cell polarity and cell migration. Adhesion receptors play a role in the control of cell proliferation and differentiation, and malfunction is involved in the development of malignancies. Adhesion receptor signaling is linked to endocytosis, gene transcription, cell cycle, apoptosis, epithelial-mesenchymal transition, tumor invasion, and tumor suppressor proteins. A variety of MAP kinase. Focal adhesions are rich in tyrosine phosphorylated proteins, coupling cell adhesion to signal transduction pathways in the cell. Various adhesion receptors, such as integrins, are closely linked to protein kinases and phosphatases. The adaptor protein Grb2 links focal adhesion kinase (FAK) to the Ras pathway when Grb2 is phosphorylated after binding to FAK. The 85 kDa subunit of the PI 3-kinase is also phosphorylated after binding to FAK. Thus, FAK is a key component in the assembly of focal contact structures that influence cytoskeletal organization and signal transduction.
Above/More Detailed Treatment: catenins : FAK : ICAMs : integrins : selectins : LEC-CAMs : adhesion : (adhesion molecules top) : cell signaling : chemotaxis : embryogenesis : ~ ERKs • GPCRs • GPCR families • hormones • Nitric Oxide• neurotransmission • neuronal interconnections ~ PKA, protein kinase A ~ PKC ~ protein kinase A ~ protein kinase C ~ protein tyrosine kinases • phosphotransfer-mediated signaling pathways • Protein Kinase Signaling Networks • receptor tyrosine kinases •• signaling gradients • signal transduction • two-component systems Џ beautiful Flash 8 animation - Inner Life of the Cell, which shows adhesion-signaling, and Interpretation: Inner Life of the Cell Џ animation MAPK signal transduction : animation G-protein :
Tables Cell Adhesion Molecules Cell signaling Immune Cytokines Second Messengers
Vascular endothelial cell adhesion and signaling during leukocyte recruitment.
During inflammation, coordinated expression of cytokine-induced adhesion molecules (CAMs) on postcapillary venular endothelial cells (ECs) regulates leukocyte recruitment. During their recruitment from blood, leukocytes adhere to EC CAMs, activating signaling pathways inside ECs. In a forthcoming paradigm, leukocyte transendothelial migration requires active EC participation, with extracellular adhesive CAM functions mirrored by cytoplasmic domain-dependent intracellular events. These events serve to reorganize the EC actin cytoskeleton. Investigators have visualized this as changes in EC shape, transient opening of EC-EC contacts, and redistribution of CAMs expressed on the luminal EC surface. In this review, we (1) summarize the overlapping extracellular adhesive properties of the 3 EC CAMs most important for leukocyte recruitment during inflammation, namely, E-selectin, vascular cell adhesion molecule, and intercellular adhesion molecule-1; (2) explore the role of these 3 CAMs as signal transducers by identifying the intracellular signals (Ca++, Rho/Rac, and phosphatidylinositol 4,5-bisphosphate) that upon leukocyte engagement, reorganize the EC cytoskeleton and redistribute these apical CAMs, thereby favoring leukocyte recruitment; and (3) describe how CAM-derived signals lead to ezrin-radixin-moesin complex formation and how this complex of plasma membrane-cytoskeleton adapter proteins coordinates CAM-driven intracellular signals with extracellular adhesive CAM functions. This literature review suggests that the cytoplasmic domains of these EC CAMs and their downstream effectors represent new and potentially beneficial intracellular therapeutic targets for treating diseases of the skin.
Kluger MS. Vascular endothelial cell adhesion and signaling during leukocyte recruitment. Adv Dermatol. 2004;20:163-201.
VCAM-1-mediated Rac signaling controls endothelial cell-cell contacts and leukocyte transmigration. [Am J Physiol Cell Physiol. 2003] PMID: 12700137
Structural pathways for macromolecular and cellular transport across the blood-brain barrier during inflammatory conditions. Review. [Histol Histopathol. 2004] PMID: 15024715
Active participation of endothelial cells in inflammation. [J Leukoc Biol. 2005] PMID: 15629883
Endothelial signaling in leukocyte transmigration. [Cell Biochem Biophys. 2003] PMID: 12794270
PECAM-1 isoform-specific activation of MAPK/ERKs and small GTPases: implications in inflammation and angiogenesis. [J Cell Biochem. 2006] PMID: 16440301
See all Related Articles...
▲ Top ▲
Tables Cell Adhesion Molecules Cell signaling Immune Cytokines Second Messengers
▼ cadherins : cadherin superfamily : catenins : cell-ECM : E-cadherin : FAK : families of CAMs : ICAM : immunoglobulin superfamily : importance of cell adhesion : integrins : selectins : signaling ▼
Cell adhesion is important in:
1. maintaining contact within solid tissue
2. embryogenesis (morphogenesis)
3. migration of single cells such as leukocytes within multicellular organisms
4. maintaining contact between neuronal elements
5. virulence of virions and bacteria
There are several families of adhesion proteins, each with specific ligands:
1. integrins with heterophilic attachments to different (hetero) ligands, including selectins with heterophilic attachments to carbohydrate ligands,
2. selectins with heterophilic attachments to carbohydrate ligands
3. Ig superfamily proteins with heterophilic attachments to: a) integrin ligands, and b) Ig superfamily proteins of a different (hetero) type, and homophilic attachments to the Ig superfamily proteins of the same (homo) type
4. cadherins with homophilic attachments to cadherins of the same type.
Cell-extracellular matrix interactions play a central role in tissue architecture and turnover. In particular, integrin-mediated cell adhesion participates in biochemical and physical signals. Cell adhesion molecules of the immunoglobulin and cadherin families, integrin heterodimers, and extracellular matrix glycoproteins, are widely expressed in the nervous system. Adhesion molecules of the immunoglobulin superfamily are involved in cell-cell adhesion, especially important during embryogenesis, wound healing, and the inflammatory response. Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion proteins. Integrins are heterodimeric molecules that function both as cell-substratum and cell-cell adhesion receptors. LEC-CAMs are cell adhesion molecules with lectin-like domains that mediate white blood cell/endothelial cell adhesion in the leukocyte adhesion cascade. Homing receptors target lymphocytes to specific lymphoid tissue.
Engagement of ICAM-1, a member of the immunoglobulin supergene family, has been documented to activate specific kinases through phosphorylation, resulting in activation of transcription factors, increased cytokine production, increased cell membrane protein expression, production of reactive oxygen species, and cell proliferation.
 Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion proteins (right - click to enlarge). The classic cadherins are defined by a conserved intracellular (i) domain which mediates interactions with cytoplasmic proteins termed catenins: α- and β-catenin. β-catenin (5) binds to both the C-terminus of the cadherin intracellular domain (6) and the N-terminus of α-catenin (4). α-catenin binds to a number of proteins involved in actin binding, bundling and polymerisation, as well as binding directly to F-actin (1) of the cytoskeleton, here through α-actin (3) in association with vinculin (2).
Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion proteins (right - click to enlarge). The classic cadherins are defined by a conserved intracellular (i) domain which mediates interactions with cytoplasmic proteins termed catenins: α- and β-catenin. β-catenin (5) binds to both the C-terminus of the cadherin intracellular domain (6) and the N-terminus of α-catenin (4). α-catenin binds to a number of proteins involved in actin binding, bundling and polymerisation, as well as binding directly to F-actin (1) of the cytoskeleton, here through α-actin (3) in association with vinculin (2).Absence of α- or β-catenin results in defective cell adhesion and failure of cadherin-catenin complexes to associate with the actin cytoskeleton.
Members of the cadherin superfamily are grouped according to the presence of one or more cadherin repeats in their extracellular (e) domains. Arrays of these (approx. 110 residue) domains form the intermolecular surfaces that mediate cadherin-mediated cell-cell interactions. Patel et al., 2003). The extracellular domains of type I and II (chordate) cadherins consist of five cadherin repeats (CRs). Type III (invertebrate and non-mammalian vertebrate) cadherins have variable numbers of CRs and also contain a region termed the primitive classical cadherin domain (PCCD) which, together with variable numbers of EGF-like and laminin G repeats, lies between the CRs and the transmembrane helix.
E-cadherin is the prototypic member of the cadherin transmembrane protein family, and regulates cell-to-cell adhesion by interacting with (homotype) E-cadherin molecules on opposing cell surfaces. E-cadherin's function in cell adhesion is also critically dependent on its ability to interact, through its cytoplasmic domain, with catenin proteins. A diverse collection of defects alter cadherin-catenin function in cancer cells.
FAK:
Some adhesion-mediated events include activation of focal adhesion kinase (FAK) with subsequent alterations in the cytoskeleton and cell morphology, changes in adhesion strength, and changes in cellular responsiveness to mechanical stimuli.[5] Fibronectin and type I collagen binding requires transcription factors (c-Fos and c-Jun) which are important in cell proliferation. A signaling pathway involving the phosphorylation of focal adhesion kinase and mitogen-activated kinases has also shown to be transiently increased in osteoblasts on fibronectin and type I collagen, but not in cells on poly-L-lysine.[6]
Ig superfamily:
The immunoglobulin superfamily is evolutionarily ancient, is widely expressed, and is constitutive or long-term up-regulated. Immunoglobulin antibodies are released by activated B cells of the immune system, on which they also act as marker, surface proteins.
Integrins:
Integrins are heterodimeric proteins so named because they integrate the function of the cell with the outside world.
Selectins:
Single transmembrane polypeptide selectins occur only in vertebrates, where they undergo calcium dependent binding via their amino-terminal domain to saccharides. The selectin family mediates function/migration of circulatory cells and the leukocyte adhesion cascade: L-leukocytes; E-endothelial cells; P-platelets and endothelial cells. The family possess large, highly glycosylated, extracellular domains, a single spanning transmembrane domain, and a small cytoplasmic tail. Selectins are rapidly downregulated by proteolytic cleavage.
Signaling molecules:
Some signaling molecules act as adhesion receptors and cluster in focal adhesions upon ligand binding. (Rho protein). Adhesion receptors promote the assembly of multi-molecular, intracellular complexes providing mechanical stability and generating intracellular signals that influence cell behavior. The functional properties of adhesion receptor complexes include roles in cell and tissue morphogenesis, cell polarity and cell migration. Adhesion receptors play a role in the control of cell proliferation and differentiation, and malfunction is involved in the development of malignancies. Adhesion receptor signaling is linked to endocytosis, gene transcription, cell cycle, apoptosis, epithelial-mesenchymal transition, tumor invasion, and tumor suppressor proteins. A variety of MAP kinase. Focal adhesions are rich in tyrosine phosphorylated proteins, coupling cell adhesion to signal transduction pathways in the cell. Various adhesion receptors, such as integrins, are closely linked to protein kinases and phosphatases. The adaptor protein Grb2 links focal adhesion kinase (FAK) to the Ras pathway when Grb2 is phosphorylated after binding to FAK. The 85 kDa subunit of the PI 3-kinase is also phosphorylated after binding to FAK. Thus, FAK is a key component in the assembly of focal contact structures that influence cytoskeletal organization and signal transduction.
Above/More Detailed Treatment: catenins : FAK : ICAMs : integrins : selectins : LEC-CAMs : adhesion : (adhesion molecules top) : cell signaling : chemotaxis : embryogenesis : ~ ERKs • GPCRs • GPCR families • hormones • Nitric Oxide• neurotransmission • neuronal interconnections ~ PKA, protein kinase A ~ PKC ~ protein kinase A ~ protein kinase C ~ protein tyrosine kinases • phosphotransfer-mediated signaling pathways • Protein Kinase Signaling Networks • receptor tyrosine kinases •• signaling gradients • signal transduction • two-component systems Џ beautiful Flash 8 animation - Inner Life of the Cell, which shows adhesion-signaling, and Interpretation: Inner Life of the Cell Џ animation MAPK signal transduction : animation G-protein :
Tables Cell Adhesion Molecules Cell signaling Immune Cytokines Second Messengers
Vascular endothelial cell adhesion and signaling during leukocyte recruitment.
During inflammation, coordinated expression of cytokine-induced adhesion molecules (CAMs) on postcapillary venular endothelial cells (ECs) regulates leukocyte recruitment. During their recruitment from blood, leukocytes adhere to EC CAMs, activating signaling pathways inside ECs. In a forthcoming paradigm, leukocyte transendothelial migration requires active EC participation, with extracellular adhesive CAM functions mirrored by cytoplasmic domain-dependent intracellular events. These events serve to reorganize the EC actin cytoskeleton. Investigators have visualized this as changes in EC shape, transient opening of EC-EC contacts, and redistribution of CAMs expressed on the luminal EC surface. In this review, we (1) summarize the overlapping extracellular adhesive properties of the 3 EC CAMs most important for leukocyte recruitment during inflammation, namely, E-selectin, vascular cell adhesion molecule, and intercellular adhesion molecule-1; (2) explore the role of these 3 CAMs as signal transducers by identifying the intracellular signals (Ca++, Rho/Rac, and phosphatidylinositol 4,5-bisphosphate) that upon leukocyte engagement, reorganize the EC cytoskeleton and redistribute these apical CAMs, thereby favoring leukocyte recruitment; and (3) describe how CAM-derived signals lead to ezrin-radixin-moesin complex formation and how this complex of plasma membrane-cytoskeleton adapter proteins coordinates CAM-driven intracellular signals with extracellular adhesive CAM functions. This literature review suggests that the cytoplasmic domains of these EC CAMs and their downstream effectors represent new and potentially beneficial intracellular therapeutic targets for treating diseases of the skin.
Kluger MS. Vascular endothelial cell adhesion and signaling during leukocyte recruitment. Adv Dermatol. 2004;20:163-201.
VCAM-1-mediated Rac signaling controls endothelial cell-cell contacts and leukocyte transmigration. [Am J Physiol Cell Physiol. 2003] PMID: 12700137
Structural pathways for macromolecular and cellular transport across the blood-brain barrier during inflammatory conditions. Review. [Histol Histopathol. 2004] PMID: 15024715
Active participation of endothelial cells in inflammation. [J Leukoc Biol. 2005] PMID: 15629883
Endothelial signaling in leukocyte transmigration. [Cell Biochem Biophys. 2003] PMID: 12794270
PECAM-1 isoform-specific activation of MAPK/ERKs and small GTPases: implications in inflammation and angiogenesis. [J Cell Biochem. 2006] PMID: 16440301
See all Related Articles...
▲ Top ▲
alternative splicing
Alternative splicing is a closely regulated, variable adaptation of the constitutive RNA modification process of pre-mRNA splicing. Alternative splicing is a form of epigenetic mechanism that enables a single gene to give rise to multiple, differentially spliced versions of a protein, increasing complexity without a change in the genome.
▼ 3'AS : 5'AS : alternative acceptor site : alternative donor site : alternative promoters : arginine-serine (RS) domain : AS-prone exons : control of AS : enhancer and repressor sequences : ESE, ESS : exon skipping : history : human genome : internal exons : introns retained : ISE, ISS : mechanisms : mutations of regulatory sequences : mutations of slice-sites : nonsense-mediated decay : protein isoforms : regulation of AS : regulatory proteins : retained introns : serine rich (SR) splicing factors : snRNP proteins : RS domain : terminal exons : tissue regulation : variable segment ▼
In 1980, a gene called 'IgM' provided the first recognized example of alternative splicing in cells—there were earlier examples in viruses. It has since been demonstrated that cells employ alternative splicing to increase protein diversity toward a wide variety of biological ends.
An average mammalian gene possesses eight or nine exons—so, since most human genes undergoing some form of alternative splicing, virtually all of these exons are candidates for elaborate control. The human genome contains 3164.7 million nucleotide bases and only around 25,000 genes, much fewer than the 90,000 proteins that we are estimated to manufacture, and much fewer than prior estimates of 80,000-140,000 expressed sequence tags (ESTs). Human Genome Project
 A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains (ORF).
A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains (ORF).
Alternative splicing can alter the mRNA product in several ways. (click to enlarge image at left)
At the simplest level, an exon can be removed (exon skip), lengthened or shortened (alternative 5'AS or 3'AS splicing). Thus, observed mechanisms of alternative splicing alteration include exon skipping, intron retention, and the use of an alternative splice donor or acceptor site. Some such alterations in gene expression cause disease.[r]
Alternative splicing can affect alternative promoters, internal exons, or alternative terminal exons, and so generates variable sequences of mRNA that shift the reading frame, insert or remove amino acids, or introduce a premature termination codon (Fig. 2). The variable segment within the mRNA results from insertion/deletion, or a mutually exclusive swap. The effects on coding potential are an in-frame insertion or deletion, a reading-frame shift, or introduction of a stop codon. mRNAs containing a stop codon >50 nt upstream of the position of the terminal intron are degraded by nonsense-mediated decay. Therefore, introduction of a premature termination codon into an mRNA by alternative splicing can be a mechanism to down-regulate expression of a gene. Alternative splicing also affects gene expression by removing or inserting regulatory elements controlling mRNA stability (NMD or nonstop decay), translation, or localization.
Exon skipping is the most frequent alternative splicing mechanism known in mammals, constituting about 40% of AS events, and as such is a major contributor to mammalian protein diversity. Exon skipping results in the loss of an exon in the alternatively spliced mRNA.
Alternative donor 5'AS or acceptor 3'AS splicing together contribute to 25% of all AS events conserved between humans and mice (4).
Intron retention is defined by the presence of a transcript-confirmed intron within a transcript-confirmed exon. Intron retention occurs when introns are not spliced out of the RNA transcript, resulting in the intron's being retained within the mRNA as part of an exon. This lengthening mechanism the commonest form of alternative splicing in plants and lower multicellular organisms.
Regulation of AS:
Alternative splicing depends upon a splice-site and nearby enhancer and repressor sequences—short segments of RNA that couple with regulatory proteins. It has been estimated that the splicing of a single exon may be promoted by at least three to seven enhancer sequences.
As a result of alternate splicing, mutations that alter a splice site or a nearby regulatory sequence can have subtle effects by shifting the ratio of the resulting proteins without entirely eliminating any form.
Regulation of alternative splicing is often temporal or tissue-specific, generating different protein isoforms in different developmental states or different tissues. At the level of the organism, specific isoforms are known to be produced as a consequence of regulation by extracellular signaling mechanisms. Alternative splicing can allow one gene to generate different proteins in different tissues, providing the diversity encoded within genomes that are smaller than would be expected from gene expression by the organism. Many highly specialized brain proteins arise from differential splicing of genes that are also expressed in other tissues. Cells can even modify splicing in response to changing conditions, and not only can alternative splicing tweak the structure of a single protein, but it may also be a means of regulating entire pathways.
Recent bioinformatics studies have demonstrated that AS-prone exons can be distinguished from constitutively spliced exons by several features(1).
1. Divisibility by three of the exon length, which is likely to ensure the preservation of the reading frame in the mRNA (3).
2. Evolutionary conservation of intronic sequences flanking AS exons (3–8). Though not yet fully understood, the unusually high conservation of the introns that surround AS exons suggests the presence of cis-regulatory elements with regulatory proteins (5).
Alternative splicing is controlled by the binding of trans-acting protein factors to cis-acting sequences within the pre-mRNA, which result in differential use of splice sites. Since the splice sites do not contain enough information to explain the complex regulation of AS, this fine-tuned mechanism is achieved by multiple weak signals across the exons and introns which are recognized by an extensive number of different proteins (2). Many such alternative-binding sequences have been identified and are grouped as either enhancer or suppressor elements. These elements are typically short (8-10 nucleotides) and are less conserved than the splice sites at exon-intron junctions. The elements are either intronic or exonic, and either enhancers or silencers depending upon whether they increase or decrease splicing – ISE, ISS, ESE, ESS.
Members of the SR (serine rich) splicing factors control alternative splice site recognition by binding to the splicing enhancer and inhibitor elements (ESE, ESS, ISE, ISS). The SR proteins interact with the pre-mRNA substrate and with snRNP proteins (small nuclear ribonucleoproteins , pronounced 'snurps'). The role of SR splicing factors in regulating splice site selection is believed to occur in arginine-serine (RS) domain dependent and RS domain independent mechanisms.
animation of alternative splicing if this link does not work then click on figure 1 of alternative splicing link : life cycle of an mRNA ~ click on Quicktime Q :HHMI Feature Article on Alternative Splicing : Artist's conception of ASControlling the Synapse — 49 Proteins at a Time : The Alternative Splicing Website : Alternative Splicing DB (ASDB) : DNA-RNA-ProteinNational Center for Biotechnology Information
Defining epigenetic states through chromatin and RNA - Nature Genetics: "The term 'epigenetics' is used to describe heritable changes in genome function that occur without a change in DNA sequence. As such, epigenetics lies at the heart of the cellular memory crucial for development and provides an important avenue for sustained response to environmental stimuli."
₪ alternative exons ₪ Alu elements ₪ alternative splicing ₪ alternative 3' splicing ₪ alternative 5' splicing ₪ cassette exons ~ cis versus trans-acting factors § domains ~ epigenetics ₪ epigenetic mechanisms ₪ ESE ₪ ESS ~ exon ₪ exon skipping ~ genes ~ gene regulation ₪ genetic variation ~ genome ₪ homeobox genes ₪ homeodomain ~ intron ₪ intron retention ₪ ISE ₪ ISS ₪ micro RNA ₪ molecular switches ~ nonsense-mediated decay ~ nonstop decay ~ nucleotide excision repair ~ open reading frame ~ pre-mRNA splicing ~ promoters ~ proteome ₪ regulation ~ regulatory proteins ф secondary antibody diversification ~ self-splicing ф signaling ₪ signaling ф somatic hypermutation, somatic mutation ~ splice-site ~ spliceosome ~ transcription ~ translation ф VDJ recombination
▲ Top ▲
▼ 3'AS : 5'AS : alternative acceptor site : alternative donor site : alternative promoters : arginine-serine (RS) domain : AS-prone exons : control of AS : enhancer and repressor sequences : ESE, ESS : exon skipping : history : human genome : internal exons : introns retained : ISE, ISS : mechanisms : mutations of regulatory sequences : mutations of slice-sites : nonsense-mediated decay : protein isoforms : regulation of AS : regulatory proteins : retained introns : serine rich (SR) splicing factors : snRNP proteins : RS domain : terminal exons : tissue regulation : variable segment ▼
In 1980, a gene called 'IgM' provided the first recognized example of alternative splicing in cells—there were earlier examples in viruses. It has since been demonstrated that cells employ alternative splicing to increase protein diversity toward a wide variety of biological ends.
An average mammalian gene possesses eight or nine exons—so, since most human genes undergoing some form of alternative splicing, virtually all of these exons are candidates for elaborate control. The human genome contains 3164.7 million nucleotide bases and only around 25,000 genes, much fewer than the 90,000 proteins that we are estimated to manufacture, and much fewer than prior estimates of 80,000-140,000 expressed sequence tags (ESTs). Human Genome Project
 A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains (ORF).
A cell typically splices a single mRNA transcript in multiple ways to generate an assortment of proteins. Alternatively spliced introns tend to lie between those exonal segments of a gene that encode the functional units, or domains (ORF).Alternative splicing can alter the mRNA product in several ways. (click to enlarge image at left)
At the simplest level, an exon can be removed (exon skip), lengthened or shortened (alternative 5'AS or 3'AS splicing). Thus, observed mechanisms of alternative splicing alteration include exon skipping, intron retention, and the use of an alternative splice donor or acceptor site. Some such alterations in gene expression cause disease.[r]
Alternative splicing can affect alternative promoters, internal exons, or alternative terminal exons, and so generates variable sequences of mRNA that shift the reading frame, insert or remove amino acids, or introduce a premature termination codon (Fig. 2). The variable segment within the mRNA results from insertion/deletion, or a mutually exclusive swap. The effects on coding potential are an in-frame insertion or deletion, a reading-frame shift, or introduction of a stop codon. mRNAs containing a stop codon >50 nt upstream of the position of the terminal intron are degraded by nonsense-mediated decay. Therefore, introduction of a premature termination codon into an mRNA by alternative splicing can be a mechanism to down-regulate expression of a gene. Alternative splicing also affects gene expression by removing or inserting regulatory elements controlling mRNA stability (NMD or nonstop decay), translation, or localization.
Exon skipping is the most frequent alternative splicing mechanism known in mammals, constituting about 40% of AS events, and as such is a major contributor to mammalian protein diversity. Exon skipping results in the loss of an exon in the alternatively spliced mRNA.
Alternative donor 5'AS or acceptor 3'AS splicing together contribute to 25% of all AS events conserved between humans and mice (4).
Intron retention is defined by the presence of a transcript-confirmed intron within a transcript-confirmed exon. Intron retention occurs when introns are not spliced out of the RNA transcript, resulting in the intron's being retained within the mRNA as part of an exon. This lengthening mechanism the commonest form of alternative splicing in plants and lower multicellular organisms.
Regulation of AS:
Alternative splicing depends upon a splice-site and nearby enhancer and repressor sequences—short segments of RNA that couple with regulatory proteins. It has been estimated that the splicing of a single exon may be promoted by at least three to seven enhancer sequences.
As a result of alternate splicing, mutations that alter a splice site or a nearby regulatory sequence can have subtle effects by shifting the ratio of the resulting proteins without entirely eliminating any form.
Regulation of alternative splicing is often temporal or tissue-specific, generating different protein isoforms in different developmental states or different tissues. At the level of the organism, specific isoforms are known to be produced as a consequence of regulation by extracellular signaling mechanisms. Alternative splicing can allow one gene to generate different proteins in different tissues, providing the diversity encoded within genomes that are smaller than would be expected from gene expression by the organism. Many highly specialized brain proteins arise from differential splicing of genes that are also expressed in other tissues. Cells can even modify splicing in response to changing conditions, and not only can alternative splicing tweak the structure of a single protein, but it may also be a means of regulating entire pathways.
Recent bioinformatics studies have demonstrated that AS-prone exons can be distinguished from constitutively spliced exons by several features(1).
1. Divisibility by three of the exon length, which is likely to ensure the preservation of the reading frame in the mRNA (3).
2. Evolutionary conservation of intronic sequences flanking AS exons (3–8). Though not yet fully understood, the unusually high conservation of the introns that surround AS exons suggests the presence of cis-regulatory elements with regulatory proteins (5).
Alternative splicing is controlled by the binding of trans-acting protein factors to cis-acting sequences within the pre-mRNA, which result in differential use of splice sites. Since the splice sites do not contain enough information to explain the complex regulation of AS, this fine-tuned mechanism is achieved by multiple weak signals across the exons and introns which are recognized by an extensive number of different proteins (2). Many such alternative-binding sequences have been identified and are grouped as either enhancer or suppressor elements. These elements are typically short (8-10 nucleotides) and are less conserved than the splice sites at exon-intron junctions. The elements are either intronic or exonic, and either enhancers or silencers depending upon whether they increase or decrease splicing – ISE, ISS, ESE, ESS.
Members of the SR (serine rich) splicing factors control alternative splice site recognition by binding to the splicing enhancer and inhibitor elements (ESE, ESS, ISE, ISS). The SR proteins interact with the pre-mRNA substrate and with snRNP proteins (small nuclear ribonucleoproteins , pronounced 'snurps'). The role of SR splicing factors in regulating splice site selection is believed to occur in arginine-serine (RS) domain dependent and RS domain independent mechanisms.
animation of alternative splicing if this link does not work then click on figure 1 of alternative splicing link : life cycle of an mRNA ~ click on Quicktime Q :HHMI Feature Article on Alternative Splicing : Artist's conception of ASControlling the Synapse — 49 Proteins at a Time : The Alternative Splicing Website : Alternative Splicing DB (ASDB) : DNA-RNA-ProteinNational Center for Biotechnology Information
Defining epigenetic states through chromatin and RNA - Nature Genetics: "The term 'epigenetics' is used to describe heritable changes in genome function that occur without a change in DNA sequence. As such, epigenetics lies at the heart of the cellular memory crucial for development and provides an important avenue for sustained response to environmental stimuli."
₪ alternative exons ₪ Alu elements ₪ alternative splicing ₪ alternative 3' splicing ₪ alternative 5' splicing ₪ cassette exons ~ cis versus trans-acting factors § domains ~ epigenetics ₪ epigenetic mechanisms ₪ ESE ₪ ESS ~ exon ₪ exon skipping ~ genes ~ gene regulation ₪ genetic variation ~ genome ₪ homeobox genes ₪ homeodomain ~ intron ₪ intron retention ₪ ISE ₪ ISS ₪ micro RNA ₪ molecular switches ~ nonsense-mediated decay ~ nonstop decay ~ nucleotide excision repair ~ open reading frame ~ pre-mRNA splicing ~ promoters ~ proteome ₪ regulation ~ regulatory proteins ф secondary antibody diversification ~ self-splicing ф signaling ₪ signaling ф somatic hypermutation, somatic mutation ~ splice-site ~ spliceosome ~ transcription ~ translation ф VDJ recombination
▲ Top ▲
allele
An allele is an alternate form of a gene at a particular location (locus) on a chromosome, and provides for genetic variability. In diploid organisms, which have two copies of each chromosome, each allele is inherited from one parent through sexual reproduction.
Alleles determine the genotype of individuals, and through genetic expression affect the phenotype of each individual. Biological evolution involves an alteration in the intergenerational frequency of alleles in populations.
Alleles determine the genotype of individuals, and through genetic expression affect the phenotype of each individual. Biological evolution involves an alteration in the intergenerational frequency of alleles in populations.
atheism
Although many scientists are religious, often deistic, the greatest proportion of atheistic scientists are to be found among biologists.

A-Deistic
Adeistic
Black Out
cosmic
Eine Kleine Nattermusing
eMusings
eVolition
Galaria
Godborygmi
Godspell Follies
Gray Matters
MetaThoughts
Mimble Wimble
Naturalism
BLogodaedaly
palimpsest
Salient
Science of Evolution
Sechuam
Sintheist
Tabula Flexuosa
The Scarlet A
Weltschauung

A-Deistic
Adeistic
Black Out
cosmic
Eine Kleine Nattermusing
eMusings
eVolition
Galaria
Godborygmi
Godspell Follies
Gray Matters
MetaThoughts
Mimble Wimble
Naturalism
BLogodaedaly
palimpsest
Salient
Science of Evolution
Sechuam
Sintheist
Tabula Flexuosa
The Scarlet A
Weltschauung
ATPases
ATP synthase is the final enzyme of oxidative phosporylation. It is located in the mitochondrial membrane and utilizes the potential energy generated by an electron transfer chain to phosphorylate ADP to ATP.
Harvard University provides a wonderful video explaining operation of the F1-F0 ATPase.
Harvard University provides a wonderful video explaining operation of the F1-F0 ATPase.
12/23/2007
base
Bases (nucleobases) are heterocyclic purine and pyrimidine analogues found in DNA & RNA: adenine / cytosine / guanine / thymine /uracil /
Nucleosides comprise the RNA bases bound to a ribose sugar molecule – examples are cytidine, uridine, adenosine, guanosine, thymidine and inosine. Deoxyribonucleosides comprise bases bound to a molecule of the sugar deoxyribose.
Nucleotides are nucleosides attached to a phosphate (phosphorylated by specific kinases), and deoxynucleotides are phosphorylated deoxyribonucleosides: AMP / ATP / cAMP / CDP / GDP / GMP / GTP / UDP / UMP /
Nucleosides comprise the RNA bases bound to a ribose sugar molecule – examples are cytidine, uridine, adenosine, guanosine, thymidine and inosine. Deoxyribonucleosides comprise bases bound to a molecule of the sugar deoxyribose.
Nucleotides are nucleosides attached to a phosphate (phosphorylated by specific kinases), and deoxynucleotides are phosphorylated deoxyribonucleosides: AMP / ATP / cAMP / CDP / GDP / GMP / GTP / UDP / UMP /
base excision repair
Base excision repair (BER) describes one form of excision repair in which damaged bases or incorrect bases are excised and replaced through the action of specific enzymes that differ between species. However, the biochemical processes involved in BER are equivalent across species, so bacterial DNA repair functions can operate in eukaryotic cells, and vice versa.
Damage is typically the result of deamination, alkylation, hydroxylation, or attack by an oxygen free radical, while the incorrect base can be uracil substituted for thymine. • C to T • C to U • T U mismatch •
In secondary antibody diversification within the adaptive immune response, activation-induced (cytidine) deaminase (AID) is involved in the initiation of three distinct immunoglobulin diversification processes: somatic hypermutation (SHM), class-switch recombination (CSR), and gene-conversion (GC). AID-catalyzed deamination of deoxycytidine creates a single nucleotide polymorphism (SNP) in the DNA strand by generating a uracil:guanine mismatch. A high-fidelity base excision repair enzyme, uracil-DNA glycosylase (UNG2), excises the alien uracil nucleobase, then error-prone DNA polymerases complete the base-excision repair. During this base-excision repair, incorrect nucleobases may be substituted at or adjacent to the original C to U mutation site. Mispairing (transition) mutations are susceptible to indels - insertions and deletions. (Such mutation vulnerable areas in the genome are termed 'hotspots', and they have played a significant role in biological evolution.)
Oxidative DNA lesions induced by oxygen free radicals such as superoxide and hydroxyl radicals appear to be repaired predominantly by base excision repair mechanisms. Further, BER is the major DNA repair system involved in removal of various oxidative DNA lesions induced by ionizing radiation - these include abasic sites and modified DNA base and sugar residues.
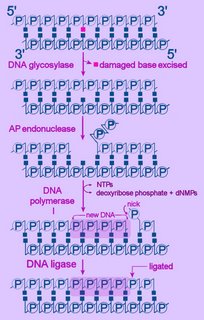 Left - diagram of base excision repair - click to enlarge image.
Left - diagram of base excision repair - click to enlarge image.
NTPs = ribonucleoside triphosphates
dNMP = deoxyribonucleoside monophosphate
First, the altered base is excised by a specific DNA glycosylase, which breaks the beta N-glycosidic bond and creates an AP, or abasic site. This site is identical to that generated by spontaneous depyrimidination or depurination. Six DNA glycosylases have been identified in humans – each excises an overlapping subset of either spontaneously formed (such as hypoxanthine), oxidized (such as 8-oxo-7,8-dihydroguanine), alkylated (such as 3-methyladenine), or mismatched (for example, T:G) bases.
Next, the terminal sugar-phosphate is removed by an AP endonuclease (Ape1), leaving a 3’-OH terminal and an abnormal 5'-abasic terminus. The resulting gap is refilled by the 5’-deoxyribose-phosphodiesterase action of a DNA polymerase I (DNA polymerase beta in mammals), then the strands are re-ligated by DNA Ligase I or a complex of XRCC1 and LigIII.
An alternative BER pathway corrects errors involving more than one nucleotide. The Fen1 protein excises the long-patch structure that is produced by DNA polymerase strand displacement. This "long-patch" repair process is divided into two subpathways: a PCNA-stimulated, Pol-beta-directed pathway and a PCNA-dependent, Pol-delta/epsilon -directed pathway.
Also: DNA repair : exogenous nucleobase rescue of abasic substitutions : link to table - human DNA repair genes : diagram>BER
▲ Top ▲
Damage is typically the result of deamination, alkylation, hydroxylation, or attack by an oxygen free radical, while the incorrect base can be uracil substituted for thymine. • C to T • C to U • T U mismatch •
In secondary antibody diversification within the adaptive immune response, activation-induced (cytidine) deaminase (AID) is involved in the initiation of three distinct immunoglobulin diversification processes: somatic hypermutation (SHM), class-switch recombination (CSR), and gene-conversion (GC). AID-catalyzed deamination of deoxycytidine creates a single nucleotide polymorphism (SNP) in the DNA strand by generating a uracil:guanine mismatch. A high-fidelity base excision repair enzyme, uracil-DNA glycosylase (UNG2), excises the alien uracil nucleobase, then error-prone DNA polymerases complete the base-excision repair. During this base-excision repair, incorrect nucleobases may be substituted at or adjacent to the original C to U mutation site. Mispairing (transition) mutations are susceptible to indels - insertions and deletions. (Such mutation vulnerable areas in the genome are termed 'hotspots', and they have played a significant role in biological evolution.)
Oxidative DNA lesions induced by oxygen free radicals such as superoxide and hydroxyl radicals appear to be repaired predominantly by base excision repair mechanisms. Further, BER is the major DNA repair system involved in removal of various oxidative DNA lesions induced by ionizing radiation - these include abasic sites and modified DNA base and sugar residues.
 Left - diagram of base excision repair - click to enlarge image.
Left - diagram of base excision repair - click to enlarge image.NTPs = ribonucleoside triphosphates
dNMP = deoxyribonucleoside monophosphate
First, the altered base is excised by a specific DNA glycosylase, which breaks the beta N-glycosidic bond and creates an AP, or abasic site. This site is identical to that generated by spontaneous depyrimidination or depurination. Six DNA glycosylases have been identified in humans – each excises an overlapping subset of either spontaneously formed (such as hypoxanthine), oxidized (such as 8-oxo-7,8-dihydroguanine), alkylated (such as 3-methyladenine), or mismatched (for example, T:G) bases.
Next, the terminal sugar-phosphate is removed by an AP endonuclease (Ape1), leaving a 3’-OH terminal and an abnormal 5'-abasic terminus. The resulting gap is refilled by the 5’-deoxyribose-phosphodiesterase action of a DNA polymerase I (DNA polymerase beta in mammals), then the strands are re-ligated by DNA Ligase I or a complex of XRCC1 and LigIII.
An alternative BER pathway corrects errors involving more than one nucleotide. The Fen1 protein excises the long-patch structure that is produced by DNA polymerase strand displacement. This "long-patch" repair process is divided into two subpathways: a PCNA-stimulated, Pol-beta-directed pathway and a PCNA-dependent, Pol-delta/epsilon -directed pathway.
Also: DNA repair : exogenous nucleobase rescue of abasic substitutions : link to table - human DNA repair genes : diagram>BER
▲ Top ▲
12/22/2007
cadherins
 Cadherins - Calcium Dependent Adhesion Molecules:
Cadherins - Calcium Dependent Adhesion Molecules: Cadherins are developmentally regulated, calcium-dependent homophilic cell-cell adhesion molecules (CAMs), through which cell-cell and cell-matrix interactions are mediated.[]im[]
The classic cadherins are defined by a conserved intracellular (i) domain which mediates interactions with cytoplasmic proteins termed catenins: α- and β-catenin. β-catenin (5) binds to both the C-terminus of the cadherin intracellular domain (6) and the N-terminus of α-catenin (4). α-catenin binds to a number of proteins involved in actin binding, bundling and polymerisation, as well as binding directly to F-actin (1) of the cytoskeleton, here through α-actin (3) in association with vinculin (2).
Absence of α- or β-catenin results in defective cell adhesion and failure of cadherin-catenin complexes to associate with the actin cytoskeleton. β-catenin is the common target for signal transduction pathways mediated by the EGF and proto-oncogenes src and Wnt-1. Several cytoplasmic Wnt regulators (β-catenin, Bcl-9/Lgs, APC, Axin) appear transiently in the nucleus.
At intercellular adherens junctions, β-catenin is an integral component of E-cadherin complexes. β-Catenin also recruits chromatin remodeling complexes, activating transcription in the nucleus. When the Wnt signaling pathway is inactive, the cytoplasmic pool of β-catenin is degraded by a complex including casein kinase 1 (CK1), glycogen synthase kinase 3 (GSK), APC and Axin, which phosphorylate β-catenin (PPPP). The phosphorylated β-catenin is recognized by the ubiquitin ligase component β-TrCP, which together with Skp1, Cul1 and the E1 and E2 ubiquitination components, mediates the ubiquitylation of β-catenin, directing it to degradation by the 26S proteasome.)[s] A third protein, p120 catenin (not shown), binds to the classic cadherin intracellular domain at a site distinct from β-catenin. Classic cadherins together with the three catenins (α-, β-, p120) form a core functional unit, the cadherin-catenin complex (CCC), which is a major component of the apical junctions formed between epithelial cells.
Members of the cadherin superfamily are grouped according to the presence of one or more cadherin repeats in their extracellular (e) domains. Arrays of these (approx. 110 residue) domains form the intermolecular surfaces that mediate cadherin-mediated cell-cell interactions. Patel et al., 2003). The extracellular domains of type I and II (chordate) cadherins consist of five cadherin repeats (CRs). Type III (invertebrate and non-mammalian vertebrate) cadherins have variable numbers of CRs and also contain a region termed the primitive classical cadherin domain (PCCD) which, together with variable numbers of EGF-like and laminin G repeats, lies between the CRs and the transmembrane helix.
E-cadherin is the prototypic member of the cadherin transmembrane protein family, and regulates cell-to-cell adhesion by interacting with (homotype) E-cadherin molecules on opposing cell surfaces. E-cadherin's function in cell adhesion is also critically dependent on its ability to interact, through its cytoplasmic domain, with catenin proteins. A diverse collection of defects alter cadherin-catenin function in cancer cells. [1, 2] The E-cadherin/catenin complex serves cell-cell adhesion and also transduces signals to the nucleus and to the cytoskeleton, either directly or through its connections with multiple other complexes. [3] The E-cadherin/catenin defects associated with malignancy include inactivating mutations and defects in the expression of E-cadherin and certain catenins, such as α-catenin.[a] Frequent inactivating mutations have been identified for the E-cadherin gene (CDH1) in diffuse gastric cancers and lobular breast cancers.[b] Members of several families of adhesion molecules appear to be important in the progression of ovarian carcinoma, including CD44, integrins, and E-cadherin. CD44 and the beta 1 integrin subunit play fundamental roles in the adhesion and migration of ovarian carcinoma cells to mesothelial cells and to their associated pericellular matrix. [4] . micro- cadherin 11 in chick embryo .
· Cadherins · calcium ion · cellular adhesion · cytokines · focal adhesion kinases · Immunoglobulins · Integrins · Rho GTPases · second messengers · Selectins · · signal transduction · two-component systems · Tables Cell Adhesion Molecules Cell signaling Immune Cytokines Second Messengers Regulatory Proteins Sequences
▲ Top ▲