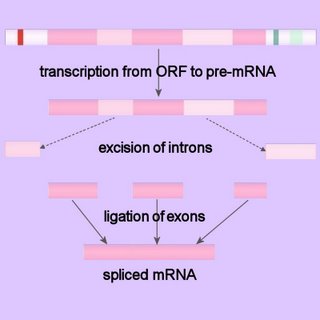
 Because eukaryotic pre-mRNAs are transcribed from intron containing genes, the sequences encoded by the intronic DNA must be removed from the primary transcript prior to the RNA's becoming biologically active. The process of intron removal is called RNA splicing, or pre-mRNA splicing. The intron-exon junctions (splice-sites) in the precursor mRNA (pre-mRNA) of eukaryotes are recognized by trans-acting factors (prokaryotes RNAs are mostly polycistronic). In pre-mRNA splicing the intronic sequences are excised and the exons are ligated to generate the spliced mRNA.
Because eukaryotic pre-mRNAs are transcribed from intron containing genes, the sequences encoded by the intronic DNA must be removed from the primary transcript prior to the RNA's becoming biologically active. The process of intron removal is called RNA splicing, or pre-mRNA splicing. The intron-exon junctions (splice-sites) in the precursor mRNA (pre-mRNA) of eukaryotes are recognized by trans-acting factors (prokaryotes RNAs are mostly polycistronic). In pre-mRNA splicing the intronic sequences are excised and the exons are ligated to generate the spliced mRNA.Group I introns occur in nuclear, mitochondrial and chloroplast rRNA genes, group II introns in mitochondrial and chloroplast mRNA genes.
Many of the group I and group II introns are self-splicing in that no additional protein factors are necessary for the intron to be efficiently and accurately excised and the strands reattached. "The nucleotide sequence of group II self-splicing introns is highly conserved, and hence these introns fold into an evolutionarily conserved three-dimensional structure, which can undergo a self-splicing reaction in the absence of any trans-acting factors [2–4]."[s]
In contrast, the nucleotide sequences and length of nuclear pre-mRNA introns is highly variable, except for the short conserved sequences at the 5´ and 3´ splice sites and the branch points*. Therefore nuclear pre-mRNA splicing requires trans-acting factors, which interact with these short conserved sequences, and from which the catalytically active spliceosome is assembled.
The conserved sequences are: 5' splice site = AGguragu; 3' splice site = yyyyyyy nagG (y= pyrimidine); branch site = ynyuray (r = purine, n = nucleotide)
Expressed differently, the highly conserved, consensus sequence for the 5' donor splice site is (for RNA): (A or C)AG/GUAAGU. That is, most exons end with AG and introns begin with GU (GT for DNA, image). The highly conserved, consensus sequence for the 3' acceptor splice site is (for RNA): (C/U)less than 10N(C/T)AG/G, where most introns end in AG after a long stretch of pyrimidines. The branch site within introns (area of lariat formation close to the acceptor site during splicing) has the consensus sequence UAUAAC (image). In most cases, U can be replaced by C and A can be replaced by G. However, the penultimate (bold) A residue is fully conserved (invariant).
Group I introns require an external guanosine nucleotide as a cofactor. The 3'-OH of the guanosine nucleotide acts as a nucleophile to attack the 5'-phosphate of the intron's 5' nucleotide. The 3' end of the 5' exon is termed the splice donor site. The 3'-OH at the 3' splice donor end of the 5' exon next attacks the splice acceptor site at the 5' nucleotide of the 3' exon, releasing the intron and covalently attaching the two exons together.
Pre-mRNA processing takes place in the nucleus of eukaryotes, whereas lack of a nuclear membrane in prokaryotes permits initiation of translation while transcription is not yet complete.
Pre-mRNA processing events include capping of the 5’ end on the pre-mRNA, pre-mRNA splicing to remove intronic sequences, and polyadenylation of the 3’ end of the pre-mRNA.
*followed by the polypyrimidine tract in metazoan introns
animation - spliceosome function : animation - spliceosome molecular action : animation of RNA splicing requires Flash Player plugin - Download plugin: clickable slide show - spliceosome intron removal :More in NCBI Molecular Cell Biology on-line text.
No comments:
Post a Comment