 Promoters direct specialized enzymes to the location at which to commence reading a segment of DNA that codes for production of a protein.
Promoters direct specialized enzymes to the location at which to commence reading a segment of DNA that codes for production of a protein.Promoters can be divided into two broad categories: those without and those with CpG islands — stretches of DNA containing multiple copies of the dinucleotide CpG, which consists of the nucleotide cytosine (C) followed by guanine (G).
In adults, single promoters with CpG islands tend to be linked to “housekeeping” genes, whereas single promoters without CpG islands are more often associated with highly regulated biological systems such as the immune and digestive systems.
A greater than expected percentage of mammalian genes have alternative promoters, which are more active during embryological development. Roughly 40-50 % of human and mouse genes have alternative promoters.
Alternative promoters can produce the same protein as single promoters, yet can be active in different tissues or at different times. In other cases, alternative promoters direct the polymerase enzymes to commence reading DNA at different start codons, ultimately resulting in different proteins with different functions. Alternative promoters, which confer greater flexibility, are more stable than single promoters over evolutionary time. The higher evolutionary conservation of alternative promoters reflects the higher density of functional elements involved in regulating promoter choice.
Alternative promoters are tightly regulated, in line with their importance in cellular function. Cells with more than one promoter regulate which promoter to use, and when. Alternative promoters are more active during embryonic development.
Promoters are DNA sequences that are recognized and bound by a DNA-dependent RNA polymerase during the initiation of transcription.
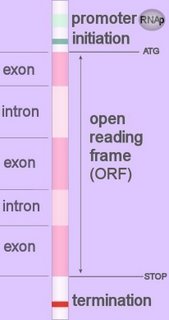
Eukaryotic promoters are located upstream (~30-100 bp, 5’) of the coding region of a gene, so they act in cis in relation to the open reading frame (ORF). Image at left - click to enlarge.
The promoter sequences:
1. Provide a variety of binding sites where RNA polymerase (RNAp), transcription factors, and other regulators of transcription bind to DNA.
2. Regulate the location and timing of transcription from the regulated gene.
3. Possess different combinations of factor binding sites depending upon how those transcripton of the ORFs is regulated.
In general, these sites can be relocated or inverted without loss of promoter activity. So, promoter sites are necessary in the immediate upstream region, yet neither their location nor orientation is essential for activity. (click to enlarge image)
Types of promoter:
(1) TATA box with a clear consensus sequence (more below).
(2) Initiators with little overall sequence conservation – initiator sequences are extremely degenerate. Like the TATA box, the initiator element positions polymerases to initiate transcription at a well-defined site.
(3) CpG islands are are CG (cytosine and guanine) rich sequences of usually 20-50 nucleotides located about 100 bp upstream from the start site. For CpG islands, initiation does not start at only one site, but rather can start at different sites within 20-200 bp. The products are RNAs with the same coding sequence, but with different 5' ends and 5' untranslated regions. The C before the G is a relatively frequent site of DNA methylation, which reduces transcriptional activity.
Several proteins termed general transcription factors are necessary for RNA polymerase II binding to chromatin templates: TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH. Highly conserved sequences within the promoter include the TATA box of eukaryotes – this is an A-T rich sequence contained in promoters for RNA polymerase II. The segment is seven base pairs long and the nucleotides most commonly found are TATAAAA.
TFIID is a complex of a TATA box binding protein (TBP) plus several proteins designated TATA binding protein-associated factors (TAFs). The TFII basal factors, especially TFIID, are analogous to bacterial sigma factors, which associate with the core RNA polymerase, and are required for correct initiation of transcription in bacteria.
A pyrimidine-rich consensus sequence – Inr – is located at or near the transcription start site. Some promoters for RNA polymerase II transcription do not contain TATA boxes, and some genes utilize an Inr site without a TATA box sequence. Some genes have a ‘downstream promoter element’ sequence important for initiation at about +30 of the transcription start site. See also Vitamin D Response Element and Serum Response Element.
The unique core promoters display a surprising diversity and that presumably reflects the diversity of interactions between promoter DNA and the proteins of the transcription complex. Promoter regions may contain response element sequences. Multiple promoters often control a single gene in parallel, adding another layer to an already complex genetic regulation mechanism. There are numerous examples in the research literature on widely separated alternative promoters that are responsible for tissue specific expression. Alternative first exons can reside in genomic locations that are far apart from each other, leading to distinct usage of proximal promoters. First exons are often poorly characterized in eukaryotic genomes.
Eubacterial promoter structure : two conserved boxes at - 10 (TATAAT) and - 35 (TTGACA) from transcription start site.
Archaeal promoter structure : TATA box and/or initiator element.
Eukaryotic promoter structure : TATA box and/or initiator element
Table Comparisons of Eubacteria, Archaea, and Eukaryotes :
image - gene control regions : 10 Genes Important for Genetic Studies : Article ~ algorithm for eukaryotic promoter recognition : Eukaryotic promoter database :
No comments:
Post a Comment