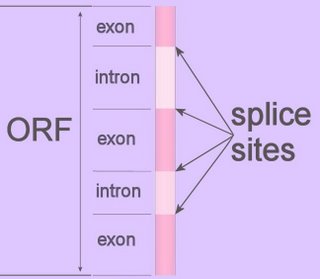
 Splice-sites are the intron-exon junctions in the precursor mRNA of eukaryotes, and are recognized by trans-acting factors (prokaryotic RNAs are mostly polycistronic).
Splice-sites are the intron-exon junctions in the precursor mRNA of eukaryotes, and are recognized by trans-acting factors (prokaryotic RNAs are mostly polycistronic).The spliceosome is a ribonuclear complex of proteins and RNA that controls genetic splicing by removing non-coding introns from the primary transcript, precursor mRNA (pre-mRNA or hnRNA). Spliceosome recognize some splice sites more readily than others, ordinarily ignoring nearby weaker sites in favor of stronger sites.
When the rate of transcription is reduced, weak sites might be spliced before stronger sites are transcribed, producing alternatively spliced protein isoforms. Cells adjust transcription rates to modulate the quantity of proteins produced, so for genes with splicing that is coupled to transcription rate, reduced transcription rates could alter not just quantity but structure of protein produced. This effect could explain why splicing is often coordinated between distant regions of the same gene–in that it's common for the inclusion of one exon to relate directly to the inclusion of another. This could mean that the splicing of each region depends rate of transcription. A slower rate of transcription could result in inclusion of exons that are excluded at a faster rate.[s, 2]
In constitutive nuclear pre-mRNA splicing the intronic sequences are excised and the exons are ligated to generate the spliced mRNA. Pre-mRNA splicing is a form of RNA processing, which yields a mature mRNA comprising the coding exons that direct sequencing of amino acids inserted into elongating polypeptides during cytoplasmic translation at ribosomes.
(click to enlarge image of splice sites)
The length and nucleotide sequence of nuclear pre-mRNA introns is highly variable, except for the short conserved sequences at the 5´ and 3´ splice sites and the branch points. Thus, splice sites immediately surrounding the intron-exon junction can be regarded as consensus sequences.
Most introns start from the sequence GU and end with the sequence AG (in the 5' donor to 3' acceptor direction). However, the sequences at the two sites are not sufficient to signal the presence of an intron, and another important sequence is called the branch site located 20 - 50 bases upstream of the acceptor site. Expressed differently, the highly conserved, consensus sequence for the 5' donor splice site is (for RNA): (A or C)AG/GUAAGU. That is, most exons end with AG and introns begin with GU (GT for DNA, diagram below left, image). The highly conserved, consensus sequence for the 3' acceptor splice site is (for RNA): (C/U)less than 10N(C/T)AG/G, where most introns end in AG after a long stretch of pyrimidines.
The branch site within introns (area of lariat formation close to the acceptor site during splicing) has the consensus sequence UAUAAC (image). In most cases, U can be replaced by C and A can be replaced by G. However, the penultimate (bold) A residue is fully conserved (invariant). Alternatively, the consensus sequence of the branch site can be expressed as "CU(A/G)A(C/U)", where A is conserved in all genes.
 Left: diagram of highly conserved, consensus (DNA) sequences for 5' donor splice site, branch site, and 3' acceptor splice site (click to enlarge).
Left: diagram of highly conserved, consensus (DNA) sequences for 5' donor splice site, branch site, and 3' acceptor splice site (click to enlarge).In over 60% of cases, the exon sequence is (A/C)AG at the donor site, and G at the acceptor site. The only splice-site feature that is 100% conserved at intron/exon junctions is that of introns beginning with GU and terminating in AG. There are, however, nucleotides that are found more frequently at particular positions (diagram below right, percentages). Vertebrates typically have a pyrimidine rich sequence (12Py) close to the 3' end of the intron. Deletion analysis has shown that although intron size varies widely, only 30-40 nucleotides at each end of an intron are required for its efficient removal.[s]
 Right: diagram of percentage occurrence of nucleotides at 5' donor splice site, branch site, and 3' acceptor splice site – rounded to 10s (click to enlarge).
Right: diagram of percentage occurrence of nucleotides at 5' donor splice site, branch site, and 3' acceptor splice site – rounded to 10s (click to enlarge).The conserved sequences can also be expressed as:
5' splice site = AGguragu
3' splice site = yyyyyyy nagG (y= pyrimidine)
branch site = ynyuray (r = purine, n = nucleotide)
alternative splicing : alternative 3' splicing : alternative 5' splicing : epigenetic mechanisms : exon skipping : intron retention
Modified: "The spliceosome is a macromolecular machine that carries out the excision of introns from eukaryotic pre-mRNAs and splicing together of exons. Four large RNA–protein complexes, called the U1, U2, U4/U6 and U5 small nuclear ribonucleoprotein particles (snRNPs), and some non-snRNP proteins assemble around three short conserved (above) sequences within the intron in an ordered manner to form the active spliceosome." [s]
"Human introns are typically thousands of bases long, and it has been reported that, in the hprt gene, sequences that match splice site consensuses (pseudosites) are highly abundant in intronic regions and that pseudoexons (i.e. intronic sequences displaying good 3' and 5' splice sites) outnumber real exons by an order of magnitude (13). Detailed analysis of one of these pseudoexons indicated that it was affected by multiple splicing defects that prevented its inclusion in the transcript. Nonetheless, other observations (14,15) suggest that a subpopulation of pseudoexons might exist in the human genome requiring only subtle changes to become splicing competent. Indeed, two recent reports (14,15) indicated that single base pair mutations or microdeletions deep within intronic regions could determine novel exon definition without creating novel splice sites, but rather altering pseudoexon sequences." Silencer elements as possible inhibitors of pseudoexon splicing Manuela Sironi, Giorgia Menozzi, Laura Riva, Rachele Cagliani, Giacomo P. Comi, Nereo Bresolin, Roberto Giorda and Uberto Pozzoli. Nucleic Acids Research, 2004, Vol. 32, No. 5 1783-1791
1 comment:
A consensus sequence may be a short sequence of nucleotides that is found several times in the genome and is thought to play the same role in its different locations.
In general, a consensus sequence is that idealized sequence in which each position represents the base/amino acid most often found when many sequences are compared. A genetic consensus sequence is a sequence of nucleotides that is common to different genes or genomes. There may be some variations but such sequences show considerable similarity. So, a consensus sequence is the prototype sequence that most others approach.
Post a Comment
Note: Only a member of this blog may post a comment.