Zinc fingers vary widely in structure and in functions, which range from DNA or RNA binding to protein-protein interactions, protein-lipid interactions, and membrane association. Zinc-finger proteins regulate the expression of genes as well as nucleic acid recognition, reverse transcription and virus assembly. In the first class of zinc fingers to be characterized, the first pair of zinc coordinating residues are cysteines, while the second pair are histidines, so the protein is termed C2H2. Other Cx ZnF classes are C4 and C6.
The FYVE zinc finger domain is conserved from yeast to man, and functions in the membrane recruitment of cytosolic proteins by binding to phosphatidylinositol 3-phosphate (PI3P), which is found mainly on endosomes [2]. The plant homeodomain (PHD) zinc finger domain has a C4HC3-type motif, and is widely distributed in eukaryotes, being found in many chromatin regulatory factors [3].[s] The RING-finger is a specialised type of Zn-finger of 40 to 60 residues that binds two atoms of zinc, and is probably involved in mediating protein-protein interactions. [2, 3, 4] E3 ubiquitin-conjugating enzymes (Ubc's) [5, 6, 7].[s].
Because they have the ability to bind to both RNA and DNA, it has been suggested that the zinc finger may represent the original nucleic acid binding protein. Zinc fingers are constituents of many regulatory proteins, many transcription factors, and receptors for steroid hormones. It has also been suggested that a Zn-centred domain could be used in a protein interaction, such as with protein kinase C. Some primary neuron-specific transcriptional regulators involved in mediating early neural development exhibit zinc finger-based DNA binding.
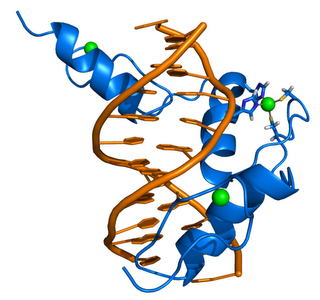 ZIF268 (Krox-24) is a transcription factor. Right - click to enlarge - Cartoon representation of a complex between DNA and the ZIF268 protein, containing 3 zinc finger motifs. The coordinating residues of the middle zinc finger are highlighted. Based on the x-ray structure of PDB 1A1L. Color coding: ZIF268: blue; DNA: orange; Zinc ions: green. Based on atomic coordinates of PDB 1A1L, rendered with open source molecular visualization tool PyMol (http://www.pymol.org/). Author: Thomas Splettstoesser
ZIF268 (Krox-24) is a transcription factor. Right - click to enlarge - Cartoon representation of a complex between DNA and the ZIF268 protein, containing 3 zinc finger motifs. The coordinating residues of the middle zinc finger are highlighted. Based on the x-ray structure of PDB 1A1L. Color coding: ZIF268: blue; DNA: orange; Zinc ions: green. Based on atomic coordinates of PDB 1A1L, rendered with open source molecular visualization tool PyMol (http://www.pymol.org/). Author: Thomas SplettstoesserDownload high-resolution version (1188x1114, 410 KB)
[] image [] MDL [] 3D steroid receptor - Zn finger []
ZnF_C2H2: Zinc finger domain comprises 25 to 30 amino-acid residues, including two conserved Cys and two conserved His residues in a C-2-C-12-H-3-H type motif. It is characterized by the sequence CX2-4C....HX2-4H, where C = cysteine, H = histidine, X = any amino acid. The 12 residues separating the second Cys and the first His are typically polar and basic amino acids, implicating this region in nucleic acid binding. ZnF C4 : consensus sequence is CX2CX13CX2CX14-15CX5CX9CX2C. The first four cysteine residues bind to a zinc ion and the last four cysteine residues bind to a second zinc ion. ZnF C6 has the consensus sequence CX2CX6CX5-6CX2CX6C. The yeast's Gal4 contains a motif where six cysteine residues interact with two zinc ions
Zinc finger projections usually interact with the major groove of the DNA double helix. The zinc fingers associate with the DNA strand such that the α-helix of each finger forms an almost continuous stretch of α-helices around the DNA molecule.
Through variations in sequence composition for the fingertip and in the number and spacing of tandem repeats of the motif, zinc fingers can form a large number of different sequence specific binding sites. Short stretches of α-helical amino acids provide binding specificity for 3 to 5 nucleobase pairs, usually short runs of guanine residues, such that the 1st, 3rd and 6th amino acid residues of the α-helix interact with the DNA helix. Amino acids at other positions can influence binding specificity by assisting binding to a specific nucleobase or by interacting with a fourth nucleobase in the complementary strand, creating target-site overlap. Where proteins contain multiple zinc fingers, each finger binds to adjacent subsites within a larger DNA recognition site, thus allowing a relatively simple motif to specifically bind to a wide range of DNA sequences.
סּ receptor proteins ~ regulatory proteins ~ transcription factors ~
▲ Top ▲
External : Tandem repeats and morphological variation
No comments:
Post a Comment