This site deals with Molecular Genetics and Biochemistry. The site is searchable using the "Search this blog" box at top left. Blue terms hyperlink to explanatory items so that you may navigate through items providing more detail. The Biochemistry Overview leads into items dealing with general biochemistry, while the Molecular Genetics Overview leads into items concerned with genetic inheritance. Use the "back" function to return to the departure item.
Items occur within Sections (listed in the sidebar). When visiting an item, the site title changes to purple – click on the title or “Home” to return to the main page.
The “Guide-Glossary” link below each item provides a glossary of terms ( # > 0 ), as a pop-up when reading within a Section, or as sub-script when visiting an Item.
12/31/2006
Biochemistry overview
Biochemistry includes the study of ions, inorganic molecules, and organic molecules involved in biological processes – for example, structural molecules, transport molecules, and metabolic pathways. Molecular genetics deals specifically with the information macromolecules involved in genetic inheritance.
12/24/2006
amines
Nitrogen is the key atom in amines. Alkyl (-CnH2n+1) or other groups are attached to the N through a carbon atom of the group. Amines are designated primary, secondary, and tertiary according to the number (1,2, or 3) of groups replacing the H atoms of an NH3 template molecule.
amino acids
Commonly called the 'building-blocks of proteins', amino acids comprise a carboxylic acid group (COOH), an attached side chain, and an amino group (NH2). The general structure of alpha amino acids is COOH-HCR-NH2, where R represents a side-chain specific to each amino acid, and the carboxyl and amino acids are attached to the same carbon atom.
Images: acidic amino acids : acidic and basic amino acids : basic amino acids : polar amino acids : non-polar amino acids :
Amino acids form peptide and protein polymers. metastream - amino acid : animation - peptide : image - primary structure protein : animation - protein : Over 90 amino acids are found in nature, while only 20 proteinogenic, standard amino acids are coded for by DNA. Of interest, 13 of the 21 amino acids found in cellular protein were generated in the Miller-Urey experiment, glycine, the smallest, being the commonest.
Several of the proteinogenic amino acids are termed ‘essential’ because they cannot be synthesized by the body’s metabolism and must be ingested in the diet. For adult humans isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine are essential, while children also require dietary histidine and arginine. Two other amino acids, selenocysteine and pyrrolysine, are sometimes incorporated during translation from RNA to protein.
In addition to their roles as substrates in peptide and protein synthesis, amino acids have other biologically important functions. Glycine, and glutamate, are both employed as neurotransmitters.
Non-standard amino acids are produced by post-translation modification, and some non-standard amino acids are produced only by plants and micro-organisms. Many amino acids are modified in the synthesis of other bio-active molecules: tryptophan is a precursor of the neurotransmitter serotonin, and glycine is one of the reactants in the synthesis of porphyrins such as heme. Numerous non-standard amino acids are also biologically important: GABA is another neurotransmitter, carnitine is employed in lipid transport within cells. Others non-standard amino acids include citrulline, homocysteine, hydroxyproline, hydroxylysine, ornithine, and sarcosine.
Virtual Cell Textbook - Biomolecules : Cell Textbook - Cell Biology :
Images: acidic amino acids : acidic and basic amino acids : basic amino acids : polar amino acids : non-polar amino acids :
Amino acids form peptide and protein polymers. metastream - amino acid : animation - peptide : image - primary structure protein : animation - protein : Over 90 amino acids are found in nature, while only 20 proteinogenic, standard amino acids are coded for by DNA. Of interest, 13 of the 21 amino acids found in cellular protein were generated in the Miller-Urey experiment, glycine, the smallest, being the commonest.
Several of the proteinogenic amino acids are termed ‘essential’ because they cannot be synthesized by the body’s metabolism and must be ingested in the diet. For adult humans isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine are essential, while children also require dietary histidine and arginine. Two other amino acids, selenocysteine and pyrrolysine, are sometimes incorporated during translation from RNA to protein.
In addition to their roles as substrates in peptide and protein synthesis, amino acids have other biologically important functions. Glycine, and glutamate, are both employed as neurotransmitters.
Non-standard amino acids are produced by post-translation modification, and some non-standard amino acids are produced only by plants and micro-organisms. Many amino acids are modified in the synthesis of other bio-active molecules: tryptophan is a precursor of the neurotransmitter serotonin, and glycine is one of the reactants in the synthesis of porphyrins such as heme. Numerous non-standard amino acids are also biologically important: GABA is another neurotransmitter, carnitine is employed in lipid transport within cells. Others non-standard amino acids include citrulline, homocysteine, hydroxyproline, hydroxylysine, ornithine, and sarcosine.
Virtual Cell Textbook - Biomolecules : Cell Textbook - Cell Biology :
amphipathic
An amphipathic, or amphiphilic molecule contains both nonpolar hydrophobic and polar hydrophilic groups.The hydrophobic group can be a long carbon chain, of the form: CH3(CH2)n, where n is greater than 4 and less than 16. Biologically important amphoteric molecules are the phospolipids, one of the main constituents of biological membranes, which naturally form bilayers. The phospolipid cell membrane insulates the cells from the surrounding medium.
12/22/2006
chemical gradients
The cell membrane is more permeable to non-polar, hydrophobic molecules than to polar, hydrophilic molecules by virtue of the hydrophobic interior of the amphipathic lipids of the bilayer. As a result, some small non-polar molecules such as H2O and CO2 are able to diffuse directly across the cell membrane down a concentration gradient. This osmotic, chemical gradient limits both the rate of diffusion and the maximum concentration of the diffusing molecule in the cytosol or the extracellular fluid (ESF) in the case of waste products.
Cells also utilize energy to generate concentration gradients across cell membranes by means of protein pumps embedded in the cell membrane. Such concentration gradients are locally discharged when ion channels are opened by a conformational change elicited by specific molecules which bind to membrane bound receptor proteins or by receptor-specific neurotransmitter molecules.
Cells also utilize energy to generate concentration gradients across cell membranes by means of protein pumps embedded in the cell membrane. Such concentration gradients are locally discharged when ion channels are opened by a conformational change elicited by specific molecules which bind to membrane bound receptor proteins or by receptor-specific neurotransmitter molecules.
cofactor
In general, a cofactor is any substance required to cooperate with an enzyme that catalyzes a specific reaction. Cofactors are non-proteinaceous substances that assist enzymes in performing catalytic actions. Cofactors may be cations (metal ions) or organic molecules known as coenzymes (vitamins).
Cofactors may be altered temporarily during the reaction which they assist, but they return to their original state after participating in catalysis, so they are not ultimately changed by the reaction.
Cofactors may be loosely or tightly bound to the enzyme. Organic, loosely bound cofactors are called coenzymes, and play an accessory role in enzyme-catalyzed processes, often by acting as a donor or acceptor of a substance involved in the reaction. When combined with an inactive apoenzyme, coenzymes form a complete, activate enzyme called the holoenzyme.
ATP and NAD+ are common coenzymes, and are loosely bound to the enzyme. Many coenzymes are phosphorylated water-soluble vitamins. Heme is tightly, covalently bound, and as such is a prosthetic group.
Prosthetic groups differ from cofactors in being tightly (often covalently) bound to a particular enzyme molecule. Haem is an example of a prosthetic group found in cytochromes and haemoglobin, which carries electrons and/or oxygen.
In molecular genetics, cofactors are regulatory proteins (activators or repressors) that interact with transcription machinery, transducing regulatory information between core RNA polymerase machinery and gene-specific transcription factors. 'Mediator' is the most universal cofactor known today – it is a modular complex serving as the interface between gene-specific RNA pol II machinery. Mediator is also needed for response to other regulatory proteins such as activators and repressors of transcription.
Index of biochemical cofactors :
Cofactors may be altered temporarily during the reaction which they assist, but they return to their original state after participating in catalysis, so they are not ultimately changed by the reaction.
Cofactors may be loosely or tightly bound to the enzyme. Organic, loosely bound cofactors are called coenzymes, and play an accessory role in enzyme-catalyzed processes, often by acting as a donor or acceptor of a substance involved in the reaction. When combined with an inactive apoenzyme, coenzymes form a complete, activate enzyme called the holoenzyme.
ATP and NAD+ are common coenzymes, and are loosely bound to the enzyme. Many coenzymes are phosphorylated water-soluble vitamins. Heme is tightly, covalently bound, and as such is a prosthetic group.
Prosthetic groups differ from cofactors in being tightly (often covalently) bound to a particular enzyme molecule. Haem is an example of a prosthetic group found in cytochromes and haemoglobin, which carries electrons and/or oxygen.
In molecular genetics, cofactors are regulatory proteins (activators or repressors) that interact with transcription machinery, transducing regulatory information between core RNA polymerase machinery and gene-specific transcription factors. 'Mediator' is the most universal cofactor known today – it is a modular complex serving as the interface between gene-specific RNA pol II machinery. Mediator is also needed for response to other regulatory proteins such as activators and repressors of transcription.
Index of biochemical cofactors :
12/20/2006
enzyme
Enzymes are organic catalysts, chemicals that increase the rate at which reaction equilibrium is achieved without themselves being permanently changed by the reaction.
Although enzymes alter k, the rate of a reaction, they do not alter Keq, the actual equilibrium point. However, if the products of a reaction are removed by a second reaction, then the reactant side of the reaction equation will be favored.
Constitutive enzymes are always produced at roughly the same concentration regardless of the composition of the medium (glycolysis, TCA cycle). Conversely, inducible enzymes are produced in response to a particular substrate, being produced only when needed. In induction, the inducer substrate, or a structurally similar compound, promotes formation of the enzyme. Conversely, the production of repressible enzymes downregulated by environmental conditions, such as the presence of the end product (corepressor) of a pathway in which the enzyme normally participates.
Regulation of enzymatic function:
Enzymes often function in metabolic chains in which regulation occurs by specific feedback mechanisms. Individual metabolic reactions are typically managed in the forward and reverse directions by structurally distinct enzymes, which permits regulation of metabolic pathways.
In bacterial cells, regulation of enzymatic reactions proceeds by:
1) control or regulation of enzyme activity by feedback inhibition or end product inhibition, which regulate biosynthetic pathways, or
2) control or regulation of synthesis of inducible or repressible enzymes, by
a) negative control as end-product repression, which downregulates enzyme synthesis and associated biosynthetic pathways,
b) positive control as enzyme induction and catabolite repression, which upregulate enzyme synthesis and associated degradative pathways
Metabolic regulation is often implemented through allosteric enzymes, which possess, as do allosteric proteins, multiple shape-changing subunits with distinct active sites. Allosteric enzymes change shape between active and inactive forms in response to the binding of substrates at the active site, or to binding of regulatory molecules at other sites. Because the reaction rates of allosteric enzymes can be regulated by only small changes in substrate concentration, allosteric enzymes are employed by cells to regulate metabolic pathways in which the concentration of cellular substrates fluctuates over narrow concentration ranges. In the simplest case in which an allosteric enzyme with a positive effector site has an active and an inactive form, the alteration in reaction rate in response to increasing substrate concentration typically displays an "S-shaped" curve. After binding of a molecule to the positive effector site of an allosteric enzyme, the second and subsequent substrates bind readily because binding of the effector molecule has induced a favorable structural change. This response is termed "cooperativity," and the S-shaped curve indicates the cooperative binding of substrate. Conversely, for allosteric enzymes with negative effector sites, binding to the allosteric site inactivates further substrate binding to the active site.
MIT Biology Hypertextbook: Enzyme Mechanisms: "Not all proteins are enzymes, but most enzymes are proteins (the exception is catalytic RNA). [Enzymes are catalysts employed in cellular reactions.] A catalyst is a molecule which increases the rate of a reaction but is not the substrate or product of that reaction. A substrate (A) is a molecule upon which an enzyme acts to yield a product (B).
A ––––> BEnz
The free energy of this reaction is not changed by the presence of the enzyme, but, for a favored reaction (where delta G is negative), the enzyme can speed it up."
Enzymes couple with substrates in transitional states, effecting conformational changes (3D structure) that facilitates transition to products.
The classification and naming of enzymes, according to the EC, depends upon their function in the reactions that they catalyze:
1. EC1. Oxidoreductases alter the oxidation state of molecules by transfer of electrons (often as hydride ions H−).
2. EC 2. Transferases transfer chemical groups from one molecule to another (not to be confused with cofactors which carry groups).
3. EC 5. Isomerases transfer chemical groups within molecules.
4. EC 3. Hydrolases add or remove H2O from molecules.
5. EC 4. Lyases manipulate double bonds in elimination reactions.
6. EC 6. Ligases condense bonds between C- and S/N/C/O, using energy from ATP
Similarly, enzymes such as RNA polymerase are named for their actions, where RNA polymerase is a common name for ATP:[DNA-directed RNA polymerase] phosphotransferase.
Specific enzymes:
DNA polymerases & reverse transcriptase : base excision repair = DNA glycosylase & AP endonuclease & Fen1 protein : hOGG1 DNA repair : helicases : RNA polymerase : ribozymes : ribozymes in repair of RNA and DNA :
Alphabetical: Enzymes →reaction→: AP endonuclease (Ape1) : aspartate aminotransferase : base excision repair : cytochrome c oxidase : DNA glycosylase : DNA Ligase I : DNA polymerases : DNA polymerase I : DNA polymerase beta : DNase IV : exonuclease 1 : exosome : Fen1 : Flap Endonuclease FEN-1 : general transcription factors : glucose-6-phosphate dehydrogenase : →glutamate-dehydrogenase→ : hOGG1 : hOGG1 oxoG oxoG repair : LigIII : MAP kinase : Msh2-Msh3 : MutS, MutL, and MutH : NADH dehydrogenase : nucleotide excision repair : 8-oxoguanine glycosylase : oxoG oxoG repair hOGG1 : PCNA : 6-phosphogluconate dehydrogenase : pyruvate carboxylase →pyruvate carboxylase→: pyruvate dehydrogenase • pyruvate dehydrogenase reaction : : RNA polymerase : replication : Replication factor C : reverse transcriptase : ribozymes : ribozymes in repair of RNA and DNA : ribulose bisphosphate carboxylase/oxygenase : RNA polymerase II : Rubisco : spliceosomal-mediated RNA trans-splicing SMaRT : succinate dehydrogenase : transaldolase : transketolase : trans-splicing ribozymes : UvrD : XRCC1 :
MIT Biology Hypertextbook on Enzyme Biochemistry : Chemical Energetics : Enzyme Mechanisms : Enzyme Kinetics : Feedback Inhibition
Although enzymes alter k, the rate of a reaction, they do not alter Keq, the actual equilibrium point. However, if the products of a reaction are removed by a second reaction, then the reactant side of the reaction equation will be favored.
Constitutive enzymes are always produced at roughly the same concentration regardless of the composition of the medium (glycolysis, TCA cycle). Conversely, inducible enzymes are produced in response to a particular substrate, being produced only when needed. In induction, the inducer substrate, or a structurally similar compound, promotes formation of the enzyme. Conversely, the production of repressible enzymes downregulated by environmental conditions, such as the presence of the end product (corepressor) of a pathway in which the enzyme normally participates.
Regulation of enzymatic function:
Enzymes often function in metabolic chains in which regulation occurs by specific feedback mechanisms. Individual metabolic reactions are typically managed in the forward and reverse directions by structurally distinct enzymes, which permits regulation of metabolic pathways.
In bacterial cells, regulation of enzymatic reactions proceeds by:
1) control or regulation of enzyme activity by feedback inhibition or end product inhibition, which regulate biosynthetic pathways, or
2) control or regulation of synthesis of inducible or repressible enzymes, by
a) negative control as end-product repression, which downregulates enzyme synthesis and associated biosynthetic pathways,
b) positive control as enzyme induction and catabolite repression, which upregulate enzyme synthesis and associated degradative pathways
Metabolic regulation is often implemented through allosteric enzymes, which possess, as do allosteric proteins, multiple shape-changing subunits with distinct active sites. Allosteric enzymes change shape between active and inactive forms in response to the binding of substrates at the active site, or to binding of regulatory molecules at other sites. Because the reaction rates of allosteric enzymes can be regulated by only small changes in substrate concentration, allosteric enzymes are employed by cells to regulate metabolic pathways in which the concentration of cellular substrates fluctuates over narrow concentration ranges. In the simplest case in which an allosteric enzyme with a positive effector site has an active and an inactive form, the alteration in reaction rate in response to increasing substrate concentration typically displays an "S-shaped" curve. After binding of a molecule to the positive effector site of an allosteric enzyme, the second and subsequent substrates bind readily because binding of the effector molecule has induced a favorable structural change. This response is termed "cooperativity," and the S-shaped curve indicates the cooperative binding of substrate. Conversely, for allosteric enzymes with negative effector sites, binding to the allosteric site inactivates further substrate binding to the active site.
MIT Biology Hypertextbook: Enzyme Mechanisms: "Not all proteins are enzymes, but most enzymes are proteins (the exception is catalytic RNA). [Enzymes are catalysts employed in cellular reactions.] A catalyst is a molecule which increases the rate of a reaction but is not the substrate or product of that reaction. A substrate (A) is a molecule upon which an enzyme acts to yield a product (B).
A ––––> BEnz
The free energy of this reaction is not changed by the presence of the enzyme, but, for a favored reaction (where delta G is negative), the enzyme can speed it up."
Enzymes couple with substrates in transitional states, effecting conformational changes (3D structure) that facilitates transition to products.
The classification and naming of enzymes, according to the EC, depends upon their function in the reactions that they catalyze:
1. EC1. Oxidoreductases alter the oxidation state of molecules by transfer of electrons (often as hydride ions H−).
2. EC 2. Transferases transfer chemical groups from one molecule to another (not to be confused with cofactors which carry groups).
3. EC 5. Isomerases transfer chemical groups within molecules.
4. EC 3. Hydrolases add or remove H2O from molecules.
5. EC 4. Lyases manipulate double bonds in elimination reactions.
6. EC 6. Ligases condense bonds between C- and S/N/C/O, using energy from ATP
Similarly, enzymes such as RNA polymerase are named for their actions, where RNA polymerase is a common name for ATP:[DNA-directed RNA polymerase] phosphotransferase.
Specific enzymes:
DNA polymerases & reverse transcriptase : base excision repair = DNA glycosylase & AP endonuclease & Fen1 protein : hOGG1 DNA repair : helicases : RNA polymerase : ribozymes : ribozymes in repair of RNA and DNA :
Alphabetical: Enzymes →reaction→: AP endonuclease (Ape1) : aspartate aminotransferase : base excision repair : cytochrome c oxidase : DNA glycosylase : DNA Ligase I : DNA polymerases : DNA polymerase I : DNA polymerase beta : DNase IV : exonuclease 1 : exosome : Fen1 : Flap Endonuclease FEN-1 : general transcription factors : glucose-6-phosphate dehydrogenase : →glutamate-dehydrogenase→ : hOGG1 : hOGG1 oxoG oxoG repair : LigIII : MAP kinase : Msh2-Msh3 : MutS, MutL, and MutH : NADH dehydrogenase : nucleotide excision repair : 8-oxoguanine glycosylase : oxoG oxoG repair hOGG1 : PCNA : 6-phosphogluconate dehydrogenase : pyruvate carboxylase →pyruvate carboxylase→: pyruvate dehydrogenase • pyruvate dehydrogenase reaction : : RNA polymerase : replication : Replication factor C : reverse transcriptase : ribozymes : ribozymes in repair of RNA and DNA : ribulose bisphosphate carboxylase/oxygenase : RNA polymerase II : Rubisco : spliceosomal-mediated RNA trans-splicing SMaRT : succinate dehydrogenase : transaldolase : transketolase : trans-splicing ribozymes : UvrD : XRCC1 :
MIT Biology Hypertextbook on Enzyme Biochemistry : Chemical Energetics : Enzyme Mechanisms : Enzyme Kinetics : Feedback Inhibition
12/17/2006
heterocyclic
Heterocyclic compounds contain a 5 or 6 atom ring structure such as found in benzene and aromatic hydrocarbons, but with other atoms substituting for ring carbons. Thus, atoms such as sulfur, oxygen or nitrogen comprise part of the ring. Examples are pyridine (C5H5N) and pyrimidine (C4H4N2), and the nucleic acids.
hydrogen bond
A hydrogen bond results from an attractive intermolecular force between two partial electric charges of opposite polarity. Although stronger than most other intermolecular forces, a hydrogen bond is much weaker than either an ionic bond or a covalent bond. Within macromolecules such as proteins and nucleic acids, hydrogen bonds can exist between two parts of the same molecule and constrain the molecule's 3D shape. More at: Wikipedia
hydrophilic
Hydrophilic, meaning 'water loving', is the chemical property of dissolving in, or orienting toward ionic, charged, solutions (such as extra- and intracellular fluids). Hydrophilic molecules or hydrophilic groups on amphipathic molecules can transiently bond with water (H2O) through the hydrogen bond. Such bonding is thermodynamically favorable, rendering these molecules soluble in water and in other polar solvents. Hydrophilic molecules are also termed polar molecules, and hydrophobic molecules are termed nonpolar molecules.
hydrophobic
Hydrophobic, meaning 'water avoiding', molecules exhibit orientation toward, or solution in, uncharged media (such as oils). This condition is opposite to hydrophilic. A hydrophobic molecule, or the hydrophobic moiety of an amphipathic molecule is typically uncharged and is not capable of hydrogen bonding. Its lack of an electrical charge enables it to dissolve more readily in oil or other hydrophobic, nonpolar solvents than in water or polar media. Hydrophilic molecules are also termed polar molecules, and hydrophobic molecules are termed nonpolar molecules.
12/09/2006
peptide
Peptides comprise linked amino acids in specific sequences. Peptides differ from proteins, which are chains of hundreds of amino acids, in being less than 50 amino acids in length. Three large classes of peptides are recognized: ribosomal peptides, nonribosomal peptides, and digested peptides.
Ribosomal peptide sequences are genetically coded, and they are synthesized at the endoplasmic reticulum by translation of mRNA. Often proteolysis generates the mature form. Ribosomal peptides function as hormones and signalling molecules.
Digested peptides are the result of nonspecific proteolysis as part of the digestive cycle.
Nonribosomal peptides are confined primarily to unicellular organisms, plants, and fungi. They are synthesized using a modular enzyme complex.
Ribosomal peptide sequences are genetically coded, and they are synthesized at the endoplasmic reticulum by translation of mRNA. Often proteolysis generates the mature form. Ribosomal peptides function as hormones and signalling molecules.
Digested peptides are the result of nonspecific proteolysis as part of the digestive cycle.
Nonribosomal peptides are confined primarily to unicellular organisms, plants, and fungi. They are synthesized using a modular enzyme complex.
basic amino acid : zwitterion with distributed charge : two amino acids joined by peptide bond
phospolipid
 Phospholipids are formed from four components: fatty acids, a negatively charged phosphate group, an alcohol and a backbone. Phospholipids with a glycerol backbone are known as glycerophospholipids or phosphoglycerides. (left - click to enlarge image)
Phospholipids are formed from four components: fatty acids, a negatively charged phosphate group, an alcohol and a backbone. Phospholipids with a glycerol backbone are known as glycerophospholipids or phosphoglycerides. (left - click to enlarge image)Essential fatty acids are required in the diet because, lacking the necessary desaturase enzymes, humans are unable to biosynthesize omega fatty acids, though we do possess the bio-machinery for their interconversion. The two closely related families of EFAs are : omega-3 (ω-3, or n-3) α-linolenic acid (18:3), and omega-6 (ω-6, n-6) linoleic acid (18:2). The EFAs serve as substrates for the biosynthesis of longer, more desaturated fatty acids (long-chain polyunsaturates).
Only sphingomyelin has a sphingosine backbone. Sphingomyelin is present in all eukaryotic cell membranes, but is mainly present in cells of the nervous system. Phospholipids, along with glycolipids and cholesterol, are a major component of all biological membranes.
tags [Biochemistry] [Molecular Biology] [eicosanoid] [phospholipid]
proteins
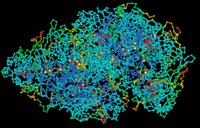 Proteins are complex, macromolecules comprised of amino acids linked by peptide bonds into long chains. The sequence (primary structure) and properties of constituent amino acids generate the 3D conformational structure (tertiary and quaternary structure) that is vital to the biological function of proteins. (click to enlarge image)
Proteins are complex, macromolecules comprised of amino acids linked by peptide bonds into long chains. The sequence (primary structure) and properties of constituent amino acids generate the 3D conformational structure (tertiary and quaternary structure) that is vital to the biological function of proteins. (click to enlarge image)Proteins are essential to the structure and biological viability of all living cells and viruses. The cellular proteome is the total cellular protein under a particular set of conditions, while the complete proteome is the sum of all potential proteomes of a cell. Proteomics has become the subject of much research in cell and molecular biology.
Proteins play a number of vital roles as:
a. Enzymes or subunits of enzymes – catalyzing cellular reactions.
b. Structural or mechanical roles – structural components of tissues, components of the cytoskeleton, centrioles, cilia and flagella, microtubules, molecular motors.
c. Intracellular and intercellular signalling functions – ion channels, receptors, membrane pumps.
d. Regulatory proteins in genetic transcription, RNA processing, spliceosomes.
e. Products of immune response that aid in targetting of foreign substances and organisms.
f. Storage and transport of various ligands.
g. The source of essential amino acids.
Almost all natural proteins are encoded by DNA, which is transcribed and processed to yield mRNA, which then serves as a template for translation by ribosomes on the rough endoplasmic reticulum.
Specific proteins/types : cAMP receptor binding protein : cofactor : core histones H2A, H2B, H3, and H4 : CRE-binding protein CREB : elongation factor EF : helicases : Helicase II : heterochromatin : histone : HP1 : inducible transcription factors : LexA repressor : mCAT2 receptor : motor proteins : nucleosome : PcG proteins : PCNA : Polycomb group : proteome : RecA : regulatory proteins : repressor proteins : ribosomes : RPA : serine rich (SR) splicing factors : silencers : Ski7p : small nuclear ribonucleoproteins (snRNPs) : spliceosome : SR (serine rich) splicing factors : trans-acting factors : trithorax group (trxG) : UPF1 UPF2 : upstream transcription factors :
Enzymes : AP endonuclease (Ape1) : DNA glycosylase : DNA Ligase I : DNA polymerases : DNA polymerase I : DNA polymerase beta : DNase IV : exonuclease 1 : exosome : Fen1 : Flap Endonuclease FEN-1 : general transcription factors : hOGG1 : hOGG1 oxoG repair : LigIII : MAP kinase : Msh2-Msh3 : MutS, MutL, and MutH : 8-oxoguanine glycosylase : oxoG repair hOGG1 : PCNA : RNA polymerase: Replication factor C : reverse transcriptase : ribozymes : RNA polymerase II : spliceosomal-mediated RNA trans-splicing SMaRT : trans-splicing ribozymes : UvrD : XRCC1 :
Rediscovering Biology - Animations and Images :
12/01/2006
zinc fingers
The zinc finger is a nucleic acid-binding protein domain in which tetrahedrally coordinated Zn ions bound to conserved cysteine and histidine residues are crucial to maintaining a hydrophobic core between α strand and two antiparallel β strands of a self-folding structure. The zinc finger is one of the most abundant DNA-binding motifs.
Zinc fingers vary widely in structure and in functions, which range from DNA or RNA binding to protein-protein interactions, protein-lipid interactions, and membrane association. Zinc-finger proteins regulate the expression of genes as well as nucleic acid recognition, reverse transcription and virus assembly. In the first class of zinc fingers to be characterized, the first pair of zinc coordinating residues are cysteines, while the second pair are histidines, so the protein is termed C2H2. Other Cx ZnF classes are C4 and C6.
The FYVE zinc finger domain is conserved from yeast to man, and functions in the membrane recruitment of cytosolic proteins by binding to phosphatidylinositol 3-phosphate (PI3P), which is found mainly on endosomes [2]. The plant homeodomain (PHD) zinc finger domain has a C4HC3-type motif, and is widely distributed in eukaryotes, being found in many chromatin regulatory factors [3].[s] The RING-finger is a specialised type of Zn-finger of 40 to 60 residues that binds two atoms of zinc, and is probably involved in mediating protein-protein interactions. [2, 3, 4] E3 ubiquitin-conjugating enzymes (Ubc's) [5, 6, 7].[s].
Because they have the ability to bind to both RNA and DNA, it has been suggested that the zinc finger may represent the original nucleic acid binding protein. Zinc fingers are constituents of many regulatory proteins, many transcription factors, and receptors for steroid hormones. It has also been suggested that a Zn-centred domain could be used in a protein interaction, such as with protein kinase C. Some primary neuron-specific transcriptional regulators involved in mediating early neural development exhibit zinc finger-based DNA binding.
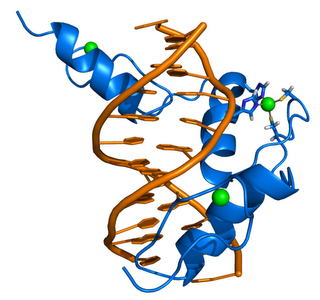 ZIF268 (Krox-24) is a transcription factor. Right - click to enlarge - Cartoon representation of a complex between DNA and the ZIF268 protein, containing 3 zinc finger motifs. The coordinating residues of the middle zinc finger are highlighted. Based on the x-ray structure of PDB 1A1L. Color coding: ZIF268: blue; DNA: orange; Zinc ions: green. Based on atomic coordinates of PDB 1A1L, rendered with open source molecular visualization tool PyMol (http://www.pymol.org/). Author: Thomas Splettstoesser
ZIF268 (Krox-24) is a transcription factor. Right - click to enlarge - Cartoon representation of a complex between DNA and the ZIF268 protein, containing 3 zinc finger motifs. The coordinating residues of the middle zinc finger are highlighted. Based on the x-ray structure of PDB 1A1L. Color coding: ZIF268: blue; DNA: orange; Zinc ions: green. Based on atomic coordinates of PDB 1A1L, rendered with open source molecular visualization tool PyMol (http://www.pymol.org/). Author: Thomas Splettstoesser
Download high-resolution version (1188x1114, 410 KB)
[] image [] MDL [] 3D steroid receptor - Zn finger []
ZnF_C2H2: Zinc finger domain comprises 25 to 30 amino-acid residues, including two conserved Cys and two conserved His residues in a C-2-C-12-H-3-H type motif. It is characterized by the sequence CX2-4C....HX2-4H, where C = cysteine, H = histidine, X = any amino acid. The 12 residues separating the second Cys and the first His are typically polar and basic amino acids, implicating this region in nucleic acid binding. ZnF C4 : consensus sequence is CX2CX13CX2CX14-15CX5CX9CX2C. The first four cysteine residues bind to a zinc ion and the last four cysteine residues bind to a second zinc ion. ZnF C6 has the consensus sequence CX2CX6CX5-6CX2CX6C. The yeast's Gal4 contains a motif where six cysteine residues interact with two zinc ions
Zinc finger projections usually interact with the major groove of the DNA double helix. The zinc fingers associate with the DNA strand such that the α-helix of each finger forms an almost continuous stretch of α-helices around the DNA molecule.
Through variations in sequence composition for the fingertip and in the number and spacing of tandem repeats of the motif, zinc fingers can form a large number of different sequence specific binding sites. Short stretches of α-helical amino acids provide binding specificity for 3 to 5 nucleobase pairs, usually short runs of guanine residues, such that the 1st, 3rd and 6th amino acid residues of the α-helix interact with the DNA helix. Amino acids at other positions can influence binding specificity by assisting binding to a specific nucleobase or by interacting with a fourth nucleobase in the complementary strand, creating target-site overlap. Where proteins contain multiple zinc fingers, each finger binds to adjacent subsites within a larger DNA recognition site, thus allowing a relatively simple motif to specifically bind to a wide range of DNA sequences.
סּ receptor proteins ~ regulatory proteins ~ transcription factors ~
▲ Top ▲
External : Tandem repeats and morphological variation
Zinc fingers vary widely in structure and in functions, which range from DNA or RNA binding to protein-protein interactions, protein-lipid interactions, and membrane association. Zinc-finger proteins regulate the expression of genes as well as nucleic acid recognition, reverse transcription and virus assembly. In the first class of zinc fingers to be characterized, the first pair of zinc coordinating residues are cysteines, while the second pair are histidines, so the protein is termed C2H2. Other Cx ZnF classes are C4 and C6.
The FYVE zinc finger domain is conserved from yeast to man, and functions in the membrane recruitment of cytosolic proteins by binding to phosphatidylinositol 3-phosphate (PI3P), which is found mainly on endosomes [2]. The plant homeodomain (PHD) zinc finger domain has a C4HC3-type motif, and is widely distributed in eukaryotes, being found in many chromatin regulatory factors [3].[s] The RING-finger is a specialised type of Zn-finger of 40 to 60 residues that binds two atoms of zinc, and is probably involved in mediating protein-protein interactions. [2, 3, 4] E3 ubiquitin-conjugating enzymes (Ubc's) [5, 6, 7].[s].
Because they have the ability to bind to both RNA and DNA, it has been suggested that the zinc finger may represent the original nucleic acid binding protein. Zinc fingers are constituents of many regulatory proteins, many transcription factors, and receptors for steroid hormones. It has also been suggested that a Zn-centred domain could be used in a protein interaction, such as with protein kinase C. Some primary neuron-specific transcriptional regulators involved in mediating early neural development exhibit zinc finger-based DNA binding.
 ZIF268 (Krox-24) is a transcription factor. Right - click to enlarge - Cartoon representation of a complex between DNA and the ZIF268 protein, containing 3 zinc finger motifs. The coordinating residues of the middle zinc finger are highlighted. Based on the x-ray structure of PDB 1A1L. Color coding: ZIF268: blue; DNA: orange; Zinc ions: green. Based on atomic coordinates of PDB 1A1L, rendered with open source molecular visualization tool PyMol (http://www.pymol.org/). Author: Thomas Splettstoesser
ZIF268 (Krox-24) is a transcription factor. Right - click to enlarge - Cartoon representation of a complex between DNA and the ZIF268 protein, containing 3 zinc finger motifs. The coordinating residues of the middle zinc finger are highlighted. Based on the x-ray structure of PDB 1A1L. Color coding: ZIF268: blue; DNA: orange; Zinc ions: green. Based on atomic coordinates of PDB 1A1L, rendered with open source molecular visualization tool PyMol (http://www.pymol.org/). Author: Thomas SplettstoesserDownload high-resolution version (1188x1114, 410 KB)
[] image [] MDL [] 3D steroid receptor - Zn finger []
ZnF_C2H2: Zinc finger domain comprises 25 to 30 amino-acid residues, including two conserved Cys and two conserved His residues in a C-2-C-12-H-3-H type motif. It is characterized by the sequence CX2-4C....HX2-4H, where C = cysteine, H = histidine, X = any amino acid. The 12 residues separating the second Cys and the first His are typically polar and basic amino acids, implicating this region in nucleic acid binding. ZnF C4 : consensus sequence is CX2CX13CX2CX14-15CX5CX9CX2C. The first four cysteine residues bind to a zinc ion and the last four cysteine residues bind to a second zinc ion. ZnF C6 has the consensus sequence CX2CX6CX5-6CX2CX6C. The yeast's Gal4 contains a motif where six cysteine residues interact with two zinc ions
Zinc finger projections usually interact with the major groove of the DNA double helix. The zinc fingers associate with the DNA strand such that the α-helix of each finger forms an almost continuous stretch of α-helices around the DNA molecule.
Through variations in sequence composition for the fingertip and in the number and spacing of tandem repeats of the motif, zinc fingers can form a large number of different sequence specific binding sites. Short stretches of α-helical amino acids provide binding specificity for 3 to 5 nucleobase pairs, usually short runs of guanine residues, such that the 1st, 3rd and 6th amino acid residues of the α-helix interact with the DNA helix. Amino acids at other positions can influence binding specificity by assisting binding to a specific nucleobase or by interacting with a fourth nucleobase in the complementary strand, creating target-site overlap. Where proteins contain multiple zinc fingers, each finger binds to adjacent subsites within a larger DNA recognition site, thus allowing a relatively simple motif to specifically bind to a wide range of DNA sequences.
סּ receptor proteins ~ regulatory proteins ~ transcription factors ~
▲ Top ▲
External : Tandem repeats and morphological variation
5/10/2006
thermochemistry
Thermochemistry refers to chemical thermodynamics – the energetic principles that underlie chemical reactions, phase changes, and solution formation. In essence, thermodynamically favourable reactions move readily from reactants to products, and are accelerated in the presence of specific catalysts or enzymes.
4/30/2006
Basics
All life is based upon chemical reactions.